An automated computational image analysis pipeline for histological grading of cardiac allograft rejection
- PMID: 33982079
- PMCID: PMC8216729
- DOI: 10.1093/eurheartj/ehab241
An automated computational image analysis pipeline for histological grading of cardiac allograft rejection
Abstract
Aim: Allograft rejection is a serious concern in heart transplant medicine. Though endomyocardial biopsy with histological grading is the diagnostic standard for rejection, poor inter-pathologist agreement creates significant clinical uncertainty. The aim of this investigation is to demonstrate that cellular rejection grades generated via computational histological analysis are on-par with those provided by expert pathologists.
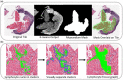
Methods and results: The study cohort consisted of 2472 endomyocardial biopsy slides originating from three major US transplant centres. The 'Computer-Assisted Cardiac Histologic Evaluation (CACHE)-Grader' pipeline was trained using an interpretable, biologically inspired, 'hand-crafted' feature extraction approach. From a menu of 154 quantitative histological features relating the density and orientation of lymphocytes, myocytes, and stroma, a model was developed to reproduce the 4-grade clinical standard for cellular rejection diagnosis. CACHE-grader interpretations were compared with independent pathologists and the 'grade of record', testing for non-inferiority (δ = 6%). Study pathologists achieved a 60.7% agreement [95% confidence interval (CI): 55.2-66.0%] with the grade of record, and pair-wise agreement among all human graders was 61.5% (95% CI: 57.0-65.8%). The CACHE-Grader met the threshold for non-inferiority, achieving a 65.9% agreement (95% CI: 63.4-68.3%) with the grade of record and a 62.6% agreement (95% CI: 60.3-64.8%) with all human graders. The CACHE-Grader demonstrated nearly identical performance in internal and external validation sets (66.1% vs. 65.8%), resilience to inter-centre variations in tissue processing/digitization, and superior sensitivity for high-grade rejection (74.4% vs. 39.5%, P < 0.001).
Conclusion: These results show that the CACHE-grader pipeline, derived using intuitive morphological features, can provide expert-quality rejection grading, performing within the range of inter-grader variability seen among human pathologists.
Keywords: Allograft rejection; Digital pathology; Heart transplant; Machine learning; Image analysis.
© The Author(s) 2021. Published by Oxford University Press on behalf of the European Society of Cardiology.
Figures




Comment in
-
Can automatic image analysis replace the pathologist in cardiac allograft rejection diagnosis?Eur Heart J. 2021 Jun 21;42(24):2370-2372. doi: 10.1093/eurheartj/ehab226. Eur Heart J. 2021. PMID: 34000014 No abstract available.
References
-
- Eisen HJ, Tuzcu EM, Dorent R, Kobashigawa J, Mancini D, Valantine-von Kaeppler HA, Starling RC, Sorensen K, Hummel M, Lind JM, Abeywickrama KH, Bernhardt P.. Everolimus for the prevention of allograft rejection and vasculopathy in cardiac-transplant recipients. N Engl J Med 2003;349:847–858. - PubMed
-
- Kobashigawa JA, Miller LW, Russell SD, Ewald GA, Zucker MJ, Goldberg LR, Eisen HJ, Salm K, Tolzman D, Gao J, Fitzsimmons W, First R.. Tacrolimus with mycophenolate mofetil (MMF) or sirolimus vs. cyclosporine with MMF in cardiac transplant patients: 1-year report. Am J Transplant 2006;6:1377–1386. - PubMed
-
- Patel JK, Kobashigawa JA.. Should we be doing routine biopsy after heart transplantation in a new era of anti-rejection? Curr Opin Cardiol 2006;21:127–131. - PubMed
-
- Costanzo MR, Dipchand A, Starling R, Anderson A, Chan M, Desai S, Fedson S, Fisher P, Gonzales-Stawinski G, Martinelli L, McGiffin D, Smith J, Taylor D, Meiser B, Webber S, Baran D, Carboni M, Dengler T, Feldman D, Frigerio M, Kfoury A, Kim D, Kobashigawa J, Shullo M, Stehlik J, Teuteberg J, Uber P, Zuckermann A, Hunt S, Burch M, Bhat G, Canter C, Chinnock R, Crespo-Leiro M, Delgado R, Dobbels F, Grady K, Kao W, Lamour J, Parry G, Patel J, Pini D, Towbin J, Wolfel G, Delgado D, Eisen H, Goldberg L, Hosenpud J, Johnson M, Keogh A, Lewis C, O'Connell J, Rogers J, Ross H, Russell S, Vanhaecke J; International Society of Heart and Lung Transplantation Guidelines. The International Society of Heart and Lung Transplantation Guidelines for the care of heart transplant recipients. J Heart Lung Transplant 2010;29:914–956. - PubMed
-
- Billingham ME, Cary NR, Hammond ME, Kemnitz J, Marboe C, McCallister HA, Snovar DC, Winters GL, Zerbe A.. A working formulation for the standardization of nomenclature in the diagnosis of heart and lung rejection: heart Rejection Study Group. The International Society for Heart Transplantation. J Heart Transplant 1990;9:587–593. - PubMed
Publication types
MeSH terms
Grants and funding
- R01 CA216579/CA/NCI NIH HHS/United States
- C06 RR012463/RR/NCRR NIH HHS/United States
- IK6 BX006185/BX/BLRD VA/United States
- I01 BX004121/BX/BLRD VA/United States
- R43 EB028736/EB/NIBIB NIH HHS/United States
- P30 CA016058/CA/NCI NIH HHS/United States
- TL1 TR001880/TR/NCATS NIH HHS/United States
- K08 HL159344/HL/NHLBI NIH HHS/United States
- R01 CA202752/CA/NCI NIH HHS/United States
- KL2 TR001879/TR/NCATS NIH HHS/United States
- R01 CA220581/CA/NCI NIH HHS/United States
- R01 HL151277/HL/NHLBI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

