Natural variation in fecundity is correlated with species-wide levels of divergence in Caenorhabditis elegans
- PMID: 33983439
- PMCID: PMC8496234
- DOI: 10.1093/g3journal/jkab168
Natural variation in fecundity is correlated with species-wide levels of divergence in Caenorhabditis elegans
Abstract
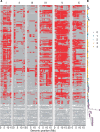
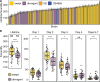
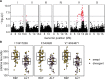
Life history traits underlie the fitness of organisms and are under strong natural selection. A new mutation that positively impacts a life history trait will likely increase in frequency and become fixed in a population (e.g., a selective sweep). The identification of the beneficial alleles that underlie selective sweeps provides insights into the mechanisms that occurred during the evolution of a species. In the global population of Caenorhabditis elegans, we previously identified selective sweeps that have drastically reduced chromosomal-scale genetic diversity in the species. Here, we measured the fecundity of 121 wild C. elegans strains, including many recently isolated divergent strains from the Hawaiian islands and found that strains with larger swept genomic regions have significantly higher fecundity than strains without evidence of the recent selective sweeps. We used genome-wide association (GWA) mapping to identify three quantitative trait loci (QTL) underlying the fecundity variation. In addition, we mapped previous fecundity data from wild C. elegans strains and C. elegans recombinant inbred advanced intercross lines that were grown in various conditions and detected eight QTL using GWA and linkage mappings. These QTL show the genetic complexity of fecundity across this species. Moreover, the haplotype structure in each GWA QTL region revealed correlations with recent selective sweeps in the C. elegans population. North American and European strains had significantly higher fecundity than most strains from Hawaii, a hypothesized origin of the C. elegans species, suggesting that beneficial alleles that caused increased fecundity could underlie the selective sweeps during the worldwide expansion of C. elegans.
Keywords: C. elegans; QTL; lifetime fecundity; natural variation; selective sweeps.
© The Author(s) 2021. Published by Oxford University Press on behalf of Genetics Society of America.
Figures


References
-
- Anderson JL, Albergotti L, Proulx S, Peden C, Huey RB, et al.2007. Thermal preference of Caenorhabditis elegans: a null model and empirical tests. J Exp Biol. 210:3107–3116. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
