Target-Based Evaluation of "Drug-Like" Properties and Ligand Efficiencies
- PMID: 33983732
- PMCID: PMC7610969
- DOI: 10.1021/acs.jmedchem.1c00416
Target-Based Evaluation of "Drug-Like" Properties and Ligand Efficiencies
Abstract
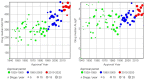
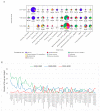
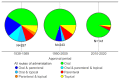
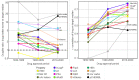
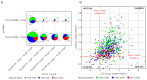
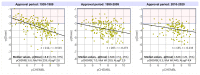
Physicochemical descriptors commonly used to define "drug-likeness" and ligand efficiency measures are assessed for their ability to differentiate marketed drugs from compounds reported to bind to their efficacious target or targets. Using ChEMBL version 26, a data set of 643 drugs acting on 271 targets was assembled, comprising 1104 drug-target pairs having ≥100 published compounds per target. Taking into account changes in their physicochemical properties over time, drugs are analyzed according to their target class, therapy area, and route of administration. Recent drugs, approved in 2010-2020, display no overall differences in molecular weight, lipophilicity, hydrogen bonding, or polar surface area from their target comparator compounds. Drugs are differentiated from target comparators by higher potency, ligand efficiency (LE), lipophilic ligand efficiency (LLE), and lower carboaromaticity. Overall, 96% of drugs have LE or LLE values, or both, greater than the median values of their target comparator compounds.
Conflict of interest statement
The authors declare no competing financial interest.
Figures



References
-
- Gleeson MP, Leeson PD, Waterbeemd Hvd. Physicochemical Properties and Compound Quality. In: Davis A, Ward SE, editors. The Handbook of Medicinal Chemistry: Principles and Practice. The Royal Society of Chemistry; 2015. pp. 1–31.
-
- Meanwell NA. Improving Drug Candidates By Design: A Focus on Physicochemical Properties as a Means of Improving Compound Disposition and Safety. Chem Res Toxicol. 2011;24:1420–1456. - PubMed
-
- Meanwell NA. Improving Drug Design: An Update on Recent Applications of Efficiency Metrics, Strategies for Replacing Problematic Elements, and Compounds In Nontraditional Drug Space. Chem Res Toxicol. 2016;29:564–616. - PubMed
-
- Young RJ. Physical Properties in Drug Design. In: Meanwell NA, editor. Tactics in Contemporary Drug Design. Springer; Berlin: 2014. pp. 1–68.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

