Integrative methylome-transcriptome analysis unravels cancer cell vulnerabilities in infant MLL-rearranged B cell acute lymphoblastic leukemia
- PMID: 33983906
- PMCID: PMC8245184
- DOI: 10.1172/JCI138833
Integrative methylome-transcriptome analysis unravels cancer cell vulnerabilities in infant MLL-rearranged B cell acute lymphoblastic leukemia
Abstract
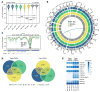
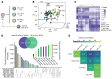
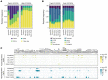
B cell acute lymphoblastic leukemia (B-ALL) is the most common childhood cancer. As predicted by its prenatal origin, infant B-ALL (iB-ALL) shows an exceptionally silent DNA mutational landscape, suggesting that alternative epigenetic mechanisms may substantially contribute to its leukemogenesis. Here, we have integrated genome-wide DNA methylome and transcriptome data from 69 patients with de novo MLL-rearranged leukemia (MLLr) and non-MLLr iB-ALL leukemia uniformly treated according to the Interfant-99/06 protocol. iB-ALL methylome signatures display a plethora of common and specific alterations associated with chromatin states related to enhancer and transcriptional control in normal hematopoietic cells. DNA methylation, gene expression, and gene coexpression network analyses segregated MLLr away from non-MLLr iB-ALL and identified a coordinated and enriched expression of the AP-1 complex members FOS and JUN and RUNX factors in MLLr iB-ALL, consistent with the significant enrichment of hypomethylated CpGs in these genes. Integrative methylome-transcriptome analysis identified consistent cancer cell vulnerabilities, revealed a robust iB-ALL-specific gene expression-correlating dmCpG signature, and confirmed an epigenetic control of AP-1 and RUNX members in reshaping the molecular network of MLLr iB-ALL. Finally, pharmacological inhibition or functional ablation of AP-1 dramatically impaired MLLr-leukemic growth in vitro and in vivo using MLLr-iB-ALL patient-derived xenografts, providing rationale for new therapeutic avenues in MLLr-iB-ALL.
Keywords: Epigenetics; Genetics; Leukemias; Oncology; Transcription.
Conflict of interest statement
Figures









Similar articles
-
Integrative epigenomic analysis identifies biomarkers and therapeutic targets in adult B-acute lymphoblastic leukemia.Cancer Discov. 2012 Nov;2(11):1004-23. doi: 10.1158/2159-8290.CD-12-0208. Epub 2012 Oct 29. Cancer Discov. 2012. PMID: 23107779 Free PMC article.
-
NG2 is a target gene of MLL-AF4 and underlies glucocorticoid resistance in MLLr B-ALL by regulating NR3C1 expression.Blood. 2024 Nov 7;144(19):2002-2017. doi: 10.1182/blood.2023022050. Blood. 2024. PMID: 39093982
-
NG2 antigen is a therapeutic target for MLL-rearranged B-cell acute lymphoblastic leukemia.Leukemia. 2019 Jul;33(7):1557-1569. doi: 10.1038/s41375-018-0353-0. Epub 2019 Jan 11. Leukemia. 2019. PMID: 30635633 Free PMC article.
-
Revisiting the biology of infant t(4;11)/MLL-AF4+ B-cell acute lymphoblastic leukemia.Blood. 2015 Dec 17;126(25):2676-85. doi: 10.1182/blood-2015-09-667378. Epub 2015 Oct 13. Blood. 2015. PMID: 26463423 Free PMC article. Review.
-
Chromatin and aberrant enhancer activity in KMT2A rearranged acute lymphoblastic leukemia.Curr Opin Genet Dev. 2024 Jun;86:102191. doi: 10.1016/j.gde.2024.102191. Epub 2024 Apr 4. Curr Opin Genet Dev. 2024. PMID: 38579381 Review.
Cited by
-
Hypoxic, glycolytic metabolism is a vulnerability of B-acute lymphoblastic leukemia-initiating cells.Cell Rep. 2022 Apr 26;39(4):110752. doi: 10.1016/j.celrep.2022.110752. Cell Rep. 2022. PMID: 35476984 Free PMC article.
-
Microfluidic-based dynamic BH3 profiling predicts anticancer treatment efficacy.NPJ Precis Oncol. 2022 Dec 1;6(1):90. doi: 10.1038/s41698-022-00333-0. NPJ Precis Oncol. 2022. PMID: 36456699 Free PMC article.
-
Mechanisms of DNA Methylation Regulatory Function and Crosstalk with Histone Lysine Methylation.J Mol Biol. 2024 Apr 1;436(7):168394. doi: 10.1016/j.jmb.2023.168394. Epub 2023 Dec 12. J Mol Biol. 2024. PMID: 38092287 Free PMC article. Review.
-
Histone acetylation by HBO1 (KAT7) activates Wnt/β-catenin signaling to promote leukemogenesis in B-cell acute lymphoblastic leukemia.Cell Death Dis. 2023 Aug 4;14(8):498. doi: 10.1038/s41419-023-06019-0. Cell Death Dis. 2023. PMID: 37542030 Free PMC article.
-
Unraveling the molecular landscape of Erdheim-Chester disease: new insights from methylome and transcriptome integration.Leukemia. 2025 Aug 21. doi: 10.1038/s41375-025-02742-z. Online ahead of print. Leukemia. 2025. PMID: 40841768
References
-
- Biondi A, et al. Biological and therapeutic aspects of infant leukemia. Blood. 2000;96(1):24–33. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

