Hierarchical semantic composition of biosimulation models using bond graphs
- PMID: 33983945
- PMCID: PMC8148364
- DOI: 10.1371/journal.pcbi.1008859
Hierarchical semantic composition of biosimulation models using bond graphs
Abstract
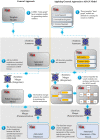
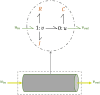
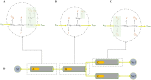
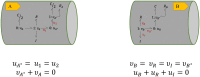
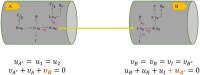
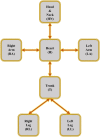
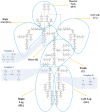
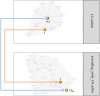
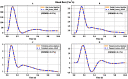
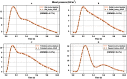
Simulating complex biological and physiological systems and predicting their behaviours under different conditions remains challenging. Breaking systems into smaller and more manageable modules can address this challenge, assisting both model development and simulation. Nevertheless, existing computational models in biology and physiology are often not modular and therefore difficult to assemble into larger models. Even when this is possible, the resulting model may not be useful due to inconsistencies either with the laws of physics or the physiological behaviour of the system. Here, we propose a general methodology for composing models, combining the energy-based bond graph approach with semantics-based annotations. This approach improves model composition and ensures that a composite model is physically plausible. As an example, we demonstrate this approach to automated model composition using a model of human arterial circulation. The major benefit is that modellers can spend more time on understanding the behaviour of complex biological and physiological systems and less time wrangling with model composition.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
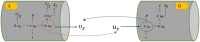

References
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

