Detection of SARS-CoV-2 RNA in bivalve mollusks and marine sediments
- PMID: 33984699
- PMCID: PMC8099584
- DOI: 10.1016/j.scitotenv.2021.147534
Detection of SARS-CoV-2 RNA in bivalve mollusks and marine sediments
Abstract
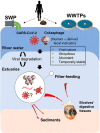
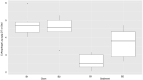
The presence of SARS-CoV-2 in wastewater pose the question of whether this new pandemic virus could be released into watercourses and potentially continue to finally reach coastal waters. In this study, we employed two bivalve molluscan species from the genus Ruditapes as sentinel organisms to investigate the presence of SARS-CoV-2 signals in the marine coastal environment. Estuarine sediments from the natural clam banks were also analyzed. Viral RNA was detected by RT-qPCR, targeting IP4, E and N1 genomic regions. Positive samples were also subjected to a PMAxx-triton viability RT-qPCR assay in order to discriminate between intact and altered capsids, obtaining indirect information about the viability of the virus. SARS-CoV-2 RNA traces were detected in 9/12 clam samples by RT-qPCR, from which 4 were positive for two different target regions. Viral quantification ranged from <LoQ to 4.48 Log genomic copies/g of digestive tissue. Regarding the sediment samples, 3/12 were positive by RT-qPCR, but only IP4 region was successfully amplificated. Quantification values for sediment samples ranged from <LoQ to 3.60 Log genomic copies/g of sediment. RNA signals disappeared in the PMAxx-triton viability RT-qPCR assay, indicating non-infectious potential. In addition, the recently discovered human-specific gut associated bacteriophage crAssphage was also quantified as a biomarker for the presence of human-derived wastewater contamination on the study area. CrAssphage was detected in 100% of both types of samples with quantification values ranging from <LoQ to 5.94 Log gc/g digestive tissue and from <LoQ to 4.71 Log gc/g sediment. Statistical analysis also showed that quantification levels for the crAssphage in clams are significantly higher than in sediments. These findings represent the first detection of SARS-CoV-2 RNA in the marine environment, demonstrating that it can reach these habitats and make contact with the marine life.
Keywords: Bivalve mollusks; COVID-19; Marine environment; SARS-CoV-2; Sediments; Wastewater.
Copyright © 2021 The Authors. Published by Elsevier B.V. All rights reserved.
Conflict of interest statement
Declaration of competing interest The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures

References
-
- Ahmed W., Angel N., Edson J., Bibby K., Bivins A., O’Brien J.W., Choi P.M., Kitajima M., Simpson S.L., Li J., Tscharke B., Verhagen R., Smith W., Zaugg J., Dierens L., Hugenholtz P., Thomas K.V., Mueller J.F. First confirmed detection of SARS-CoV-2 in untreated wastewater in Australia: a proof of concept for the wastewater surveillance of COVID-19 in the community. Sci. Total Environ. 2020;728 doi: 10.1016/j.scitotenv.2020.138764. - DOI - PMC - PubMed
-
- Butler D., Davies J.W. In: Urban Drainage. Butler D., Davies J.W., editors. Spon Press, Taylor and Francis Group; New York, USA: 2004. Combined sewers and combined sewer overflows; pp. 254–289.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

