Cyclic oligoadenylate signalling and regulation by ring nucleases during type III CRISPR defence
- PMID: 33986148
- PMCID: PMC8284326
- DOI: 10.1261/rna.078739.121
Cyclic oligoadenylate signalling and regulation by ring nucleases during type III CRISPR defence
Abstract
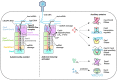
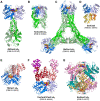
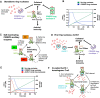
In prokaryotes, CRISPR-Cas immune systems recognise and cleave foreign nucleic acids to defend against Mobile Genetic Elements (MGEs). Type III CRISPR-Cas complexes also synthesise cyclic oligoadenylate (cOA) second messengers, which activate CRISPR ancillary proteins involved in antiviral defence. In particular, cOA-stimulated nucleases degrade RNA and DNA non-specifically, which slows MGE replication but also impedes cell growth, necessitating mechanisms to eliminate cOA in order to mitigate collateral damage. Extant cOA is degraded by a new class of enzyme termed a 'ring nuclease', which cleaves cOA specifically and switches off CRISPR ancillary enzymes. Several ring nuclease families have been characterised to date, including a family used by MGEs to circumvent CRISPR immunity, and encompass diverse protein folds and distinct cOA cleavage mechanisms. In this review we outline cOA signalling, discuss how different ring nucleases regulate the cOA signalling pathway, and reflect on parallels between cyclic nucleotide-based immune systems to reveal new areas for exploration.
Keywords: CARF; CRISPR-Cas; Csm6 ribonuclease; cyclic nucleotides; ring nuclease.
Published by Cold Spring Harbor Laboratory Press for the RNA Society.
Figures

Similar articles
-
Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.Nucleic Acids Res. 2023 Oct 27;51(19):10590-10605. doi: 10.1093/nar/gkad739. Nucleic Acids Res. 2023. PMID: 37747760 Free PMC article.
-
Tetramerisation of the CRISPR ring nuclease Crn3/Csx3 facilitates cyclic oligoadenylate cleavage.Elife. 2020 Jun 29;9:e57627. doi: 10.7554/eLife.57627. Elife. 2020. PMID: 32597755 Free PMC article.
-
Enzymatic properties of CARF-domain proteins in Synechocystis sp. PCC 6803.Front Microbiol. 2022 Nov 7;13:1046388. doi: 10.3389/fmicb.2022.1046388. eCollection 2022. Front Microbiol. 2022. PMID: 36419420 Free PMC article.
-
Type III CRISPR-Cas: beyond the Cas10 effector complex.Trends Biochem Sci. 2024 Jan;49(1):28-37. doi: 10.1016/j.tibs.2023.10.006. Epub 2023 Nov 8. Trends Biochem Sci. 2024. PMID: 37949766 Free PMC article. Review.
-
The diverse arsenal of type III CRISPR-Cas-associated CARF and SAVED effectors.Biochem Soc Trans. 2022 Oct 31;50(5):1353-1364. doi: 10.1042/BST20220289. Biochem Soc Trans. 2022. PMID: 36282000 Free PMC article. Review.
Cited by
-
The SAVED domain of the type III CRISPR protease CalpL is a ring nuclease.Nucleic Acids Res. 2024 Sep 23;52(17):10520-10532. doi: 10.1093/nar/gkae676. Nucleic Acids Res. 2024. PMID: 39166476 Free PMC article.
-
CRISPR-Cas, Argonaute proteins and the emerging landscape of amplification-free diagnostics.Methods. 2022 Sep;205:1-10. doi: 10.1016/j.ymeth.2022.06.002. Epub 2022 Jun 9. Methods. 2022. PMID: 35690249 Free PMC article.
-
The effect of crRNA-target mismatches on cOA-mediated interference by a type III-A CRISPR-Cas system.RNA Biol. 2022 Jan;19(1):1293-1304. doi: 10.1080/15476286.2022.2150812. RNA Biol. 2022. PMID: 36424814 Free PMC article.
-
Activation of Csm6 ribonuclease by cyclic nucleotide binding: in an emergency, twist to open.Nucleic Acids Res. 2023 Oct 27;51(19):10590-10605. doi: 10.1093/nar/gkad739. Nucleic Acids Res. 2023. PMID: 37747760 Free PMC article.
-
RecA-dependent or independent recombination of plasmid DNA generates a conflict with the host EcoKI immunity by launching restriction alleviation.Nucleic Acids Res. 2024 May 22;52(9):5195-5208. doi: 10.1093/nar/gkae243. Nucleic Acids Res. 2024. PMID: 38567730 Free PMC article.
References
-
- Aguirre S, Luthra P, Sanchez-Aparicio MT, Maestre AM, Patel J, Lamothe F, Fredericks AC, Tripathi S, Zhu T, Pintado-Silva J, et al. 2017. Dengue virus NS2B protein targets cGAS for degradation and prevents mitochondrial DNA sensing during infection. Nat Microbiol 2: 17037. 10.1038/nmicrobiol.2017.37 - DOI - PMC - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
