SLFN2 protection of tRNAs from stress-induced cleavage is essential for T cell-mediated immunity
- PMID: 33986151
- PMCID: PMC8442736
- DOI: 10.1126/science.aba4220
SLFN2 protection of tRNAs from stress-induced cleavage is essential for T cell-mediated immunity
Abstract
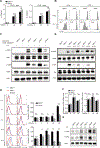
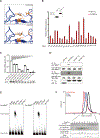
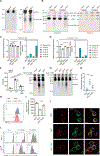
Reactive oxygen species (ROS) increase in activated T cells because of metabolic activity induced to support T cell proliferation and differentiation. We show that these ROS trigger an oxidative stress response that leads to translation repression. This response is countered by Schlafen 2 (SLFN2), which directly binds transfer RNAs (tRNAs) to protect them from cleavage by the ribonuclease angiogenin. T cell-specific SLFN2 deficiency results in the accumulation of tRNA fragments, which inhibit translation and promote stress-granule formation. Interleukin-2 receptor β (IL-2Rβ) and IL-2Rγ fail to be translationally up-regulated after T cell receptor stimulation, rendering SLFN2-deficient T cells insensitive to interleukin-2's mitogenic effects. SLFN2 confers resistance against the ROS-mediated translation-inhibitory effects of oxidative stress normally induced by T cell activation, permitting the robust protein synthesis necessary for T cell expansion and immunity.
Copyright © 2021 The Authors, some rights reserved; exclusive licensee American Association for the Advancement of Science. No claim to original U.S. Government Works.
Figures

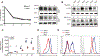

Comment in
-
De-stressing the T cells in need.Science. 2021 May 14;372(6543):683-684. doi: 10.1126/science.abi7265. Science. 2021. PMID: 33986167 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

