Repurposing potential of posaconazole and grazoprevir as inhibitors of SARS-CoV-2 helicase
- PMID: 33986405
- PMCID: PMC8119689
- DOI: 10.1038/s41598-021-89724-0
Repurposing potential of posaconazole and grazoprevir as inhibitors of SARS-CoV-2 helicase
Abstract
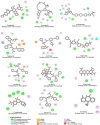
As the Severe Acute Respiratory Syndrome Coronavirus-2 (SARS-CoV-2) pandemic engulfs millions worldwide, the quest for vaccines or drugs against the virus continues. The helicase protein of SARS-CoV-2 represents an attractive target for drug discovery since inhibition of helicase activity can suppress viral replication. Using in silico approaches, we have identified drugs that interact with SARS-CoV-2 helicase based on the presence of amino acid arrangements matching binding sites of drugs in previously annotated protein structures. The drugs exhibiting an RMSD of ≤ 3.0 Å were further analyzed using molecular docking, molecular dynamics (MD) simulation, and post-MD analyses. Using these approaches, we found 12 drugs that showed strong interactions with SARS-CoV-2 helicase amino acids. The analyses were performed using the recently available SARS-CoV-2 helicase structure (PDB ID: 5RL6). Based on the MM-GBSA approach, out of the 12 drugs, two drugs, namely posaconazole and grazoprevir, showed the most favorable binding energy, - 54.8 and - 49.1 kcal/mol, respectively. Furthermore, of the amino acids found conserved among all human coronaviruses, 10/11 and 10/12 were targeted by, respectively, grazoprevir and posaconazole. These residues are part of the crucial DEAD-like helicase C and DEXXQc_Upf1-like/ DEAD-like helicase domains. Strong interactions of posaconazole and grazoprevir with conserved amino acids indicate that the drugs can be potent against SARS-CoV-2. Since the amino acids are conserved among the human coronaviruses, the virus is unlikely to develop resistance mutations against these drugs. Since these drugs are already in use, they may be immediately repurposed for SARS-CoV-2 therapy.
Conflict of interest statement
The authors declare no competing interests.
Figures






References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

