Improved Microbial Community Characterization of 16S rRNA via Metagenome Hybridization Capture Enrichment
- PMID: 33986735
- PMCID: PMC8110821
- DOI: 10.3389/fmicb.2021.644662
Improved Microbial Community Characterization of 16S rRNA via Metagenome Hybridization Capture Enrichment
Abstract
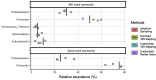
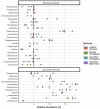
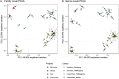
Environmental microbial diversity is often investigated from a molecular perspective using 16S ribosomal RNA (rRNA) gene amplicons and shotgun metagenomics. While amplicon methods are fast, low-cost, and have curated reference databases, they can suffer from amplification bias and are limited in genomic scope. In contrast, shotgun metagenomic methods sample more genomic regions with fewer sequence acquisition biases, but are much more expensive (even with moderate sequencing depth) and computationally challenging. Here, we develop a set of 16S rRNA sequence capture baits that offer a potential middle ground with the advantages from both approaches for investigating microbial communities. These baits cover the diversity of all 16S rRNA sequences available in the Greengenes (v. 13.5) database, with no sequence having <78% sequence identity to at least one bait for all segments of 16S. The use of our baits provide comparable results to 16S amplicon libraries and shotgun metagenomic libraries when assigning taxonomic units from 16S sequences within the metagenomic reads. We demonstrate that 16S rRNA capture baits can be used on a range of microbial samples (i.e., mock communities and rodent fecal samples) to increase the proportion of 16S rRNA sequences (average > 400-fold) and decrease analysis time to obtain consistent community assessments. Furthermore, our study reveals that bioinformatic methods used to analyze sequencing data may have a greater influence on estimates of community composition than library preparation method used, likely due in part to the extent and curation of the reference databases considered. Thus, enriching existing aliquots of shotgun metagenomic libraries and obtaining modest numbers of reads from them offers an efficient orthogonal method for assessment of bacterial community composition.
Keywords: amplicon; microbial diversity; microbiome; mock communities; next generation sequencing; shotgun libraries; target enrichment.
Copyright © 2021 Beaudry, Wang, Kieran, Thomas, Bayona-Vásquez, Gao, Devault, Brunelle, Lu, Wang, Rhodes and Glenn.
Conflict of interest statement
The EHS DNA lab provides oligonucleotide aliquots and library preparation services at cost, including some oligonucleotides and services used in this manuscript (baddna.uga.edu). BB and AD were employed by, and thereby have financial interest in, Daicel Arbor Biosciences, who provided the in-solution capture reagents used in this work. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Altschul S. F., Gish W., Miller W., Myers E., Lipman D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215 403–410. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources

