Adhesive Functions or Pseudogenization of Type Va Autotransporters in Brucella Species
- PMID: 33987105
- PMCID: PMC8111173
- DOI: 10.3389/fcimb.2021.607610
Adhesive Functions or Pseudogenization of Type Va Autotransporters in Brucella Species
Abstract
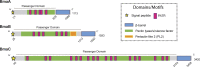
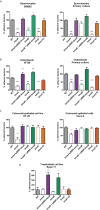
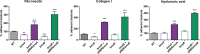
Adhesion to host cells is a key step for successful infection of many bacterial pathogens and may define tropism to different host tissues. To do so, bacteria display adhesins on their surfaces. Brucella is an intracellular pathogen capable of proliferating in a wide variety of cell types. It has been described that BmaC, a large protein that belongs to the classical (type Va) autotransporter family, is required for efficient adhesion of Brucella suis strain 1330 to epithelial cells and fibronectin. Here we show that B. suis 1330 harbors two other type Va autotransporters (BmaA and BmaB), which, although much smaller, share significant sequence similarities with BmaC and contain the essential domains to mediate proper protein translocation to the bacterial surface. Gain and loss of function studies indicated that BmaA, BmaB, and BmaC contribute, to a greater or lesser degree, to adhesion of B. suis 1330 to different cells such as synovial fibroblasts, osteoblasts, trophoblasts, and polarized epithelial cells as well as to extracellular matrix components. It was previously shown that BmaC localizes to a single bacterial pole. Interestingly, we observed here that, similar to BmaC, the BmaB adhesin is localized mostly at a single cell pole, reinforcing the hypothesis that Brucella displays an adhesive pole. Although Brucella species have strikingly similar genomes, they clearly differ in their host preferences. Mainly, the differences identified between species appear to be at loci encoding surface proteins. A careful in silico analysis of the putative type Va autotransporter orthologues from several Brucella strains showed that the bmaB locus from Brucella abortus and both, the bmaA and bmaC loci from Brucella melitensis are pseudogenes in all strains analyzed. Results reported here evidence that all three autotransporters play a role in the adhesion properties of B. suis 1330. However, Brucella spp. exhibit extensive variations in the repertoire of functional adhesins of the classical autotransporter family that can be displayed on the bacterial surface, making them an interesting target for future studies on host preference and tropism.
Keywords: Brucella; adhesin; adhesion; extracellular matrix (ECM); outer membrane protein (OMP); polar localization; pseudogene; type Va autotransporter.
Copyright © 2021 Bialer, Ferrero, Delpino, Ruiz-Ranwez, Posadas, Baldi and Zorreguieta.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
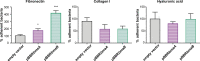


Similar articles
-
BmaC, a novel autotransporter of Brucella suis, is involved in bacterial adhesion to host cells.Cell Microbiol. 2012 Jun;14(6):965-82. doi: 10.1111/j.1462-5822.2012.01771.x. Epub 2012 Mar 8. Cell Microbiol. 2012. PMID: 22321605
-
BtaE, an adhesin that belongs to the trimeric autotransporter family, is required for full virulence and defines a specific adhesive pole of Brucella suis.Infect Immun. 2013 Mar;81(3):996-1007. doi: 10.1128/IAI.01241-12. Epub 2013 Jan 14. Infect Immun. 2013. PMID: 23319562 Free PMC article.
-
Adhesins of Brucella: Their Roles in the Interaction with the Host.Pathogens. 2020 Nov 12;9(11):942. doi: 10.3390/pathogens9110942. Pathogens. 2020. PMID: 33198223 Free PMC article. Review.
-
The BtaF trimeric autotransporter of Brucella suis is involved in attachment to various surfaces, resistance to serum and virulence.PLoS One. 2013 Nov 13;8(11):e79770. doi: 10.1371/journal.pone.0079770. eCollection 2013. PLoS One. 2013. PMID: 24236157 Free PMC article.
-
Phylogenetic Classification and Functional Review of Autotransporters.Front Immunol. 2022 Jul 1;13:921272. doi: 10.3389/fimmu.2022.921272. eCollection 2022. Front Immunol. 2022. PMID: 35860281 Free PMC article. Review.
Cited by
-
Kinetics of Placental Infection by Different Smooth Brucella Strains in Mice.Pathogens. 2022 Feb 22;11(3):279. doi: 10.3390/pathogens11030279. Pathogens. 2022. PMID: 35335603 Free PMC article.
-
Cell and Tissue Tropism of Brucella spp.Infect Immun. 2023 May 16;91(5):e0006223. doi: 10.1128/iai.00062-23. Epub 2023 Apr 27. Infect Immun. 2023. PMID: 37129522 Free PMC article. Review.
-
Loss to gain: pseudogenes in microorganisms, focusing on eubacteria, and their biological significance.Appl Microbiol Biotechnol. 2024 May 8;108(1):328. doi: 10.1007/s00253-023-12971-w. Appl Microbiol Biotechnol. 2024. PMID: 38717672 Free PMC article. Review.
-
Membrane Protein Bcest Is Involved in Hyphal Growth, Virulence and Stress Tolerance of Botrytis cinerea.Microorganisms. 2023 May 6;11(5):1225. doi: 10.3390/microorganisms11051225. Microorganisms. 2023. PMID: 37317199 Free PMC article.
-
Cross regulation in a three-component cell envelope stress signaling system of Brucella.bioRxiv [Preprint]. 2023 Oct 11:2023.04.15.536747. doi: 10.1101/2023.04.15.536747. bioRxiv. 2023. Update in: mBio. 2023 Dec 19;14(6):e0238723. doi: 10.1128/mbio.02387-23. PMID: 37873345 Free PMC article. Updated. Preprint.
References
-
- Bachvarov B., Kirilov K., Ivanov I. (2008). Codon Usage in Prokaryotes. Biotechnol. Biotechnol. Equip. 22, 669–682. 10.1080/13102818.2008.10817533 - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

