Potential functional variants of KIAA genes are associated with breast cancer risk in a case control study
- PMID: 33987247
- PMCID: PMC8105804
- DOI: 10.21037/atm-20-6108
Potential functional variants of KIAA genes are associated with breast cancer risk in a case control study
Abstract
Background: KIAA genes identified in the Kazusa cDNA-sequencing project may play important roles in biological processes and are involved in carcinogenesis of many cancers. Genetic variants of KIAA genes are implicated in the abnormal expression of these genes and are linked to susceptibility of several human complex diseases.
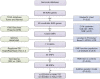
Methods: The differentially expressed KIAA genes were screened and identified in The Cancer Genome Atlas (TCGA) database of breast cancer. A total of 48 variants located in the 28 KIAA genes were selected to investigate the associations between polymorphism and breast cancer in 1,032 cases and 1,063 cancer-free controls in a Chinese population.
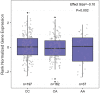
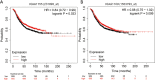
Results: Two coding variants, which included a SNP rs2306369 in KIAA1109 and a SNP rs1205434 in KIAA1755, were identified to be associated with the incidences of breast cancer. Logistic regression analysis showed that the SNP rs2306369 G allele was associated with a decreased risk of breast cancer (additive model: OR =0.81, 95% CI: 0.66-0.99, P=0.038), whereas the SNP rs1205434 A allele was involved with a higher risk of breast cancer (additive model: OR =1.19, 95% CI: 1.02-1.38, P= 0.025). Further stratified analysis revealed that the SNP rs1205434 showed a significant difference for age at menarche strata (heterogeneity test P=0.009). Multiplicative interaction analysis indicated that there was positive multiplicative interaction between the SNP rs1205434 and menarche age (OR =1.09, 95% CI: 1.01-1.17, P=0.036). Additionally, expression quantitative trait loci analysis revealed that the SNP rs1205434 A allele could decrease the KIAA1755 expression in the Genotype-Tissue Expression (GTEx) database (P=0.002). The Kaplan-Meier plotter showed that breast cancer patients with high KIAA1755 expression have significantly better outcomes than those with low levels of expression (HR =0.84, 95% CI: 0.72-0.99, P=0.033).
Conclusions: The results indicate that the genetic variants (rs2306369 and rs1205434) in the coding region of KIAA1109 and KIAA1755 respectively may affect Chinese females' breast cancer susceptibility and act as potential predictive biomarkers for breast cancer.
Keywords: KIAA; breast cancer; genetic variants; susceptibility.
2021 Annals of Translational Medicine. All rights reserved.
Conflict of interest statement
Conflicts of Interest: All authors have completed the ICMJE uniform disclosure form (available at http://dx.doi.org/10.21037/atm-20-6108). The authors declare that they have no conflict of interest regarding the publication of this paper.
Figures
Similar articles
-
Coding variants in the PCNT and CEP295 genes contribute to breast cancer risk in Chinese women.Pathol Res Pract. 2021 Sep;225:153581. doi: 10.1016/j.prp.2021.153581. Epub 2021 Aug 4. Pathol Res Pract. 2021. PMID: 34418690
-
Rare coding variants and breast cancer risk: evaluation of susceptibility Loci identified in genome-wide association studies.Cancer Epidemiol Biomarkers Prev. 2014 Apr;23(4):622-8. doi: 10.1158/1055-9965.EPI-13-1043. Epub 2014 Jan 27. Cancer Epidemiol Biomarkers Prev. 2014. PMID: 24470074 Free PMC article.
-
Evaluation of genetic variants in autophagy pathway genes as prognostic biomarkers for breast cancer.Gene. 2017 Sep 5;627:549-555. doi: 10.1016/j.gene.2017.06.053. Epub 2017 Jun 29. Gene. 2017. PMID: 28669927
-
Construction of expression-ready cDNA clones for KIAA genes: manual curation of 330 KIAA cDNA clones.DNA Res. 2002 Jun 30;9(3):99-106. doi: 10.1093/dnares/9.3.99. DNA Res. 2002. PMID: 12168954 Review.
-
Kazusa mammalian cDNA resources: towards functional characterization of KIAA gene products.Brief Funct Genomic Proteomic. 2006 Mar;5(1):4-7. doi: 10.1093/bfgp/ell005. Epub 2006 Feb 20. Brief Funct Genomic Proteomic. 2006. PMID: 16769670 Review.
Cited by
-
Targeting Hypoxia and HIF1α in Triple-Negative Breast Cancer: New Insights from Gene Expression Profiling and Implications for Therapy.Biology (Basel). 2024 Jul 31;13(8):577. doi: 10.3390/biology13080577. Biology (Basel). 2024. PMID: 39194515 Free PMC article.
-
Prognostic Value of Necroptosis-Related Genes Signature in Oral Squamous Cell Carcinoma.Cancers (Basel). 2023 Sep 13;15(18):4539. doi: 10.3390/cancers15184539. Cancers (Basel). 2023. PMID: 37760507 Free PMC article.
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
