Subgrouping by gene expression profiles to improve relapse risk prediction in paediatric B-precursor acute lymphoblastic leukaemia
- PMID: 33987975
- PMCID: PMC8178509
- DOI: 10.1002/cam4.3842
Subgrouping by gene expression profiles to improve relapse risk prediction in paediatric B-precursor acute lymphoblastic leukaemia
Abstract
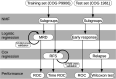
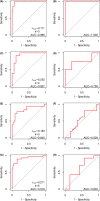
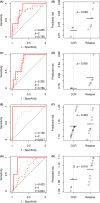
Relapsed acute lymphoblastic leukaemia (ALL) remains a prevalent paediatric cancer and one of the most common causes of mortality from malignancy in children. Tailoring the intensity of therapy according to early stratification is a promising strategy but remains a major challenge due to heterogeneity and subtyping difficulty. In this study, we subgroup B-precursor ALL patients by gene expression profiles, using non-negative matrix factorization and minimum description length which unsupervisedly determines the number of subgroups. Within each of the four subgroups, logistic and Cox regression with elastic net regularization are used to build models predicting minimal residual disease (MRD) and relapse-free survival (RFS) respectively. Measured by area under the receiver operating characteristic curve (AUC), subgrouping improves prediction of MRD in one subgroup which mostly overlaps with subtype TCF3-PBX1 (AUC = 0·986 in the training set and 1·0 in the test set), compared to a global model published previously. The models predicting RFS displayed acceptable concordance in training set and discriminate high-relapse-risk patients in three subgroups of the test set (Wilcoxon test p = 0·048, 0·036, and 0·016). Genes playing roles in the models are specific to different subgroups. The improvement of subgrouped MRD prediction and the differences of genes in prediction models of subgroups suggest that the heterogeneity of B-precursor ALL can be handled by subgrouping according to gene expression profiles to improve the prediction accuracy.
Keywords: B-precursor acute lymphoblastic leukaemia; gene expression profiles; minimal residual disease; non-negative matrix factorization; relapse.
© 2021 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures


References
-
- Hunger SP, Mullighan CG. Acute lymphoblastic leukemia in children. N Engl J Med. 2015;373(16):1541‐1552. - PubMed
-
- Holleman A, Cheok MH, den Boer ML, et al. Gene‐expression patterns in drug‐resistant acute lymphoblastic leukemia cells and response to treatment. N Engl J Med. 2004;351(6):533‐542. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources

