Ubiquitylation in DNA double-strand break repair
- PMID: 33990032
- PMCID: PMC8207832
- DOI: 10.1016/j.dnarep.2021.103129
Ubiquitylation in DNA double-strand break repair
Abstract
Genome integrity is constantly challenged by various DNA lesions with DNA double-strand breaks (DSBs) as the most cytotoxic lesions. In order to faithfully repair DSBs, DNA damage response (DDR) signaling networks have evolved, which organize many multi-protein complexes to deal with the encountered DNA damage. Spatiotemporal dynamics of these protein complexes at DSBs are mainly modulated by post-translational modifications (PTMs). One of the most well-studied PTMs in DDR is ubiquitylation which can orchestrate cellular responses to DSBs, promote accurate DNA repair, and maintain genome integrity. Here, we summarize the recent advances of ubiquitin-dependent signaling in DDR and discuss how ubiquitylation crosstalks with other PTMs to control fundamental biological processes in DSB repair.
Keywords: DNA damage response; Double strand break; homologous recombination (HR); non-homologous end joining (NHEJ); ubiquitylation.
Copyright © 2021 Elsevier B.V. All rights reserved.
Conflict of interest statement
Conflict of interest
The authors declare no conflict of interest.
Figures
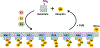
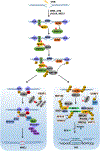

References
-
- Harper JW, Elledge SJ, The DNA damage response: ten years after, Mol Cell, 28 (2007) 739–745. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

