Knl1 participates in spindle assembly checkpoint signaling in maize
- PMID: 33990465
- PMCID: PMC8157932
- DOI: 10.1073/pnas.2022357118
Knl1 participates in spindle assembly checkpoint signaling in maize
Abstract
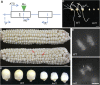
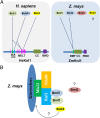
The Knl1-Mis12-Ndc80 (KMN) network is an essential component of the kinetochore-microtubule attachment interface, which is required for genomic stability in eukaryotes. However, little is known about plant Knl1 proteins because of their complex evolutionary history. Here, we cloned the Knl1 homolog from maize (Zea mays) and confirmed it as a constitutive central kinetochore component. Functional assays demonstrated their conserved role in chromosomal congression and segregation during nuclear division, thus causing defective cell division during kernel development when Knl1 transcript was depleted. A 145 aa region in the middle of maize Knl1, that did not involve the MELT repeats, was associated with the interaction of spindle assembly checkpoint (SAC) components Bub1/Mad3 family proteins 1 and 2 (Bmf1/2) but not with the Bmf3 protein. They may form a helical conformation with a hydrophobic interface with the TPR domain of Bmf1/2, which is similar to that of vertebrates. However, this region detected in monocots shows extensive divergence in eudicots, suggesting that distinct modes of the SAC to kinetochore connection are present within plant lineages. These findings elucidate the conserved role of the KMN network in cell division and a striking dynamic of evolutionary patterns in the SAC signaling and kinetochore network.
Keywords: Knl1; SAC; cell division; kinetochore; maize.
Conflict of interest statement
The authors declare no competing interest.
Figures





Similar articles
-
KI motifs of human Knl1 enhance assembly of comprehensive spindle checkpoint complexes around MELT repeats.Curr Biol. 2014 Jan 6;24(1):29-39. doi: 10.1016/j.cub.2013.11.046. Epub 2013 Dec 19. Curr Biol. 2014. PMID: 24361068
-
A coadapted KNL1 and spindle assembly checkpoint axis orchestrates precise mitosis in Arabidopsis.Proc Natl Acad Sci U S A. 2024 Jan 9;121(2):e2316583121. doi: 10.1073/pnas.2316583121. Epub 2024 Jan 3. Proc Natl Acad Sci U S A. 2024. PMID: 38170753 Free PMC article.
-
How the SAC gets the axe: Integrating kinetochore microtubule attachments with spindle assembly checkpoint signaling.Bioarchitecture. 2015;5(1-2):1-12. doi: 10.1080/19490992.2015.1090669. Epub 2015 Oct 2. Bioarchitecture. 2015. PMID: 26430805 Free PMC article. Review.
-
Arabidopsis KNL1 recruits type one protein phosphatase to kinetochores to silence the spindle assembly checkpoint.Sci Adv. 2025 Feb 7;11(6):eadq4033. doi: 10.1126/sciadv.adq4033. Epub 2025 Feb 5. Sci Adv. 2025. PMID: 39908360 Free PMC article.
-
The dynamic protein Knl1 - a kinetochore rendezvous.J Cell Sci. 2014 Aug 15;127(Pt 16):3415-23. doi: 10.1242/jcs.149922. Epub 2014 Jul 22. J Cell Sci. 2014. PMID: 25052095 Review.
Cited by
-
The Loss-Function of KNL1 Causes Oligospermia and Asthenospermia in Mice by Affecting the Assembly and Separation of the Spindle through Flow Cytometry and Immunofluorescence.Sensors (Basel). 2023 Feb 25;23(5):2571. doi: 10.3390/s23052571. Sensors (Basel). 2023. PMID: 36904774 Free PMC article.
-
Microtubule Regulation in Plants: From Morphological Development to Stress Adaptation.Biomolecules. 2023 Mar 30;13(4):627. doi: 10.3390/biom13040627. Biomolecules. 2023. PMID: 37189374 Free PMC article. Review.
-
Non-B-form DNA tends to form in centromeric regions and has undergone changes in polyploid oat subgenomes.Proc Natl Acad Sci U S A. 2023 Jan 3;120(1):e2211683120. doi: 10.1073/pnas.2211683120. Epub 2022 Dec 27. Proc Natl Acad Sci U S A. 2023. PMID: 36574697 Free PMC article.
-
Integrative mapping in large inbred and hybrid association panels along with an F2 population advanced a novel understanding of general combining ability for plant height in maize.Theor Appl Genet. 2025 Apr 1;138(4):90. doi: 10.1007/s00122-025-04883-2. Theor Appl Genet. 2025. PMID: 40169419
-
Centromeres: From chromosome biology to biotechnology applications and synthetic genomes in plants.Plant Biotechnol J. 2022 Nov;20(11):2051-2063. doi: 10.1111/pbi.13875. Epub 2022 Jul 7. Plant Biotechnol J. 2022. PMID: 35722725 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

