The endoribonuclease N4BP1 prevents psoriasis by controlling both keratinocytes proliferation and neutrophil infiltration
- PMID: 33990547
- PMCID: PMC8121926
- DOI: 10.1038/s41419-021-03774-w
The endoribonuclease N4BP1 prevents psoriasis by controlling both keratinocytes proliferation and neutrophil infiltration
Abstract
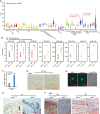
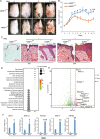
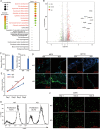
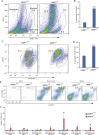
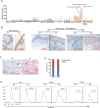
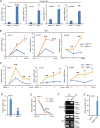
Psoriasis is a common chronic skin disease, characterized by abnormal interplay between hyperproliferative epidermal keratinocytes and self-reactive immune cells with not fully addressed molecular mechanism. N4BP1 (NEDD4-binding protein 1) is considered as an immune regulator for a long time but its physiological role is not determined yet. Here, we found that the expression of N4BP1 in skin was highest among all 54 tested tissues, and its expression was further upregulated in psoriatic skin. N4BP1-deficient mice exhibited normal grossly, but developed severe and prolonged IMQ-induced psoriasis-like disease comparing to controls. N4BP1 mainly expressed in keratinocytes and located on nucleus. Up- but not downregulated genes in N4BP1-deficient skin were specifically enriched in keratinocyte proliferation and differentiation. The proliferation of N4BP1-deficient primary keratinocytes was faster compared to that of controls. The upregulated genes upon ablation of N4BP1 were highly enriched in targets of AP-1 transcription factor. Knocking out N4BP1 resulted in upregulation of JunB and FosB, and conversely, overexpression of N4BP1 greatly reduced their expression. Furthermore, N4BP1 binds with JunB and FosB encoding mRNAs and greatly reduces their stability. In addition, with a high expression in neutrophils, N4BP1 limits survival of neutrophils in blood and infiltration of neutrophils in psoriatic skin by targeting CXCL1, CCL20, and S100A8. These findings demonstrate that N4BP1 controls the proper function of keratinocytes and neutrophils by negatively regulating JunB, FosB, and CXCL1, respectively, and that is critical for psoriasis prevention.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Research Materials
Miscellaneous

