Exploring ligand binding pathways on proteins using hypersound-accelerated molecular dynamics
- PMID: 33990583
- PMCID: PMC8121818
- DOI: 10.1038/s41467-021-23157-1
Exploring ligand binding pathways on proteins using hypersound-accelerated molecular dynamics
Abstract
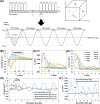
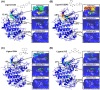
Capturing the dynamic processes of biomolecular systems in atomistic detail remains difficult despite recent experimental advances. Although molecular dynamics (MD) techniques enable atomic-level observations, simulations of "slow" biomolecular processes (with timescales longer than submilliseconds) are challenging because of current computer speed limitations. Therefore, we developed a method to accelerate MD simulations by high-frequency ultrasound perturbation. The binding events between the protein CDK2 and its small-molecule inhibitors were nearly undetectable in 100-ns conventional MD, but the method successfully accelerated their slow binding rates by up to 10-20 times. Hypersound-accelerated MD simulations revealed a variety of microscopic kinetic features of the inhibitors on the protein surface, such as the existence of different binding pathways to the active site. Moreover, the simulations allowed the estimation of the corresponding kinetic parameters and exploring other druggable pockets. This method can thus provide deeper insight into the microscopic interactions controlling biomolecular processes.
Conflict of interest statement
The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

