Biofilm viability checker: An open-source tool for automated biofilm viability analysis from confocal microscopy images
- PMID: 33990612
- PMCID: PMC8121819
- DOI: 10.1038/s41522-021-00214-7
Biofilm viability checker: An open-source tool for automated biofilm viability analysis from confocal microscopy images
Abstract
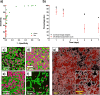
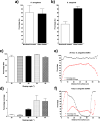
Quantifying biofilm formation on surfaces is challenging because traditional microbiological methods, such as total colony-forming units (CFUs), often rely on manual counting. These are laborious, resource intensive techniques, more susceptible to human error. Confocal laser scanning microscopy (CLSM) is a high-resolution technique that allows 3D visualisation of biofilm architecture. In combination with a live/dead stain, it can be used to quantify biofilm viability on both transparent and opaque surfaces. However, there is little consensus on the appropriate methodology to apply in confocal micrograph processing. In this study, we report the development of an image analysis approach to repeatably quantify biofilm viability and surface coverage. We also demonstrate its use for a range of bacterial species and translational applications. This protocol has been created with ease of use and accessibility in mind, to enable researchers who do not specialise in computational techniques to be confident in applying these methods to analyse biofilm micrographs. Furthermore, the simplicity of the method enables the user to adapt it for their bespoke needs. Validation experiments demonstrate the automated analysis is robust and accurate across a range of bacterial species and an improvement on traditional microbiological analysis. Furthermore, application to translational case studies show the automated method is a reliable measurement of biomass and cell viability. This approach will ensure image analysis is an accessible option for those in the microbiology and biomaterials field, improve current detection approaches and ultimately support the development of novel strategies for preventing biofilm formation by ensuring comparability across studies.
Conflict of interest statement
The authors declare no competing interests.
Figures

Similar articles
-
Introducing BAIT (Biofilm Architecture Inference Tool): a software program to evaluate the architecture of oral multi-species biofilms.Microbiology (Reading). 2019 May;165(5):527-537. doi: 10.1099/mic.0.000761. Epub 2019 Mar 18. Microbiology (Reading). 2019. PMID: 30882296
-
Enhanced analysis of bacteria susceptibility in connected biofilms.J Microbiol Methods. 2012 Jul;90(1):9-14. doi: 10.1016/j.mimet.2012.04.008. Epub 2012 Apr 19. J Microbiol Methods. 2012. PMID: 22542520
-
A two-step procedure for automatic and accurate segmentation of volumetric CLSM biofilm images.J Microbiol Methods. 2007 Sep;70(3):424-33. doi: 10.1016/j.mimet.2007.05.022. Epub 2007 Jun 16. J Microbiol Methods. 2007. PMID: 17618700
-
Critical review on biofilm methods.Crit Rev Microbiol. 2017 May;43(3):313-351. doi: 10.1080/1040841X.2016.1208146. Epub 2016 Nov 21. Crit Rev Microbiol. 2017. PMID: 27868469 Review.
-
Tools of the Trade: Image Analysis Programs for Confocal Laser-Scanning Microscopy Studies of Biofilms and Considerations for Their Use by Experimental Researchers.ACS Omega. 2023 May 26;8(23):20163-20177. doi: 10.1021/acsomega.2c07255. eCollection 2023 Jun 13. ACS Omega. 2023. PMID: 37332792 Free PMC article. Review.
Cited by
-
Poly-Gamma-Glutamic Acid Nanopolymer Effect against Bacterial Biofilms: In Vitro and In Vivo Study.Biomedicines. 2024 Jan 23;12(2):251. doi: 10.3390/biomedicines12020251. Biomedicines. 2024. PMID: 38397853 Free PMC article.
-
Effect of biofilm physical characteristics on their susceptibility to antibiotics: impacts of low-frequency ultrasound.NPJ Biofilms Microbiomes. 2024 Aug 19;10(1):70. doi: 10.1038/s41522-024-00544-2. NPJ Biofilms Microbiomes. 2024. PMID: 39160204 Free PMC article.
-
A Simple High-Throughput Technology for Microorganism Detection and Quantitative Analysis.Foods. 2024 Sep 18;13(18):2954. doi: 10.3390/foods13182954. Foods. 2024. PMID: 39335881 Free PMC article.
-
Cinacalcet exhibits rapid bactericidal and efficient anti-biofilm activities against multidrug-resistant Gram-positive pathogens.iScience. 2023 Mar 11;26(4):106378. doi: 10.1016/j.isci.2023.106378. eCollection 2023 Apr 21. iScience. 2023. PMID: 37034999 Free PMC article.
-
Eicosapentaenoic acid as an antibiofilm agent disrupts mature biofilms of Candida albicans.Biofilm. 2024 Dec 30;9:100251. doi: 10.1016/j.bioflm.2024.100251. eCollection 2025 Jun. Biofilm. 2024. PMID: 39845529 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

