Upregulation of miR-3195, miR-3687 and miR-4417 is associated with castration-resistant prostate cancer
- PMID: 33990872
- PMCID: PMC8519832
- DOI: 10.1007/s00345-021-03723-4
Upregulation of miR-3195, miR-3687 and miR-4417 is associated with castration-resistant prostate cancer
Abstract
Purpose: Prostate cancer (PCa) is a leading cause of cancer-related death. Upon androgen-deprivation therapy, the disease may progress further to castration-resistant PCa (CRPC) with a poor prognosis. MicroRNAs (miRNAs) are small non-coding RNAs, which play crucial roles in gene regulation. The aim of our study is to find CRPC-associated miRNAs and to evaluate their functional role.
Methods: In this study, 23 benign prostatic hyperplasia (BPH), 76 primary PCa, and 35 CRPC specimens were included. Total RNA extracted from tissue sections was used for miRNA profiling on the Affymetrix GSC 3000 platform. Subsequently, stem-loop RT-qPCR analysis was performed to validate the expression levels of selected miRNAs. PCa cell lines were transfected with miRNA mimics or inhibitors to evaluate the effects on cell proliferation, cell migration and cell invasion.
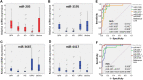
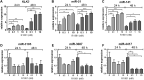
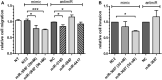
Results: In our profiling study, several miRNAs were found to be deregulated in CRPC compared to primary PCa tissue, of which miR-205 (- 4.5-fold; p = 0.0009), miR-92b (- 3.1 fold; p < 0.0001) were downregulated and miR-3195 (5.6-fold; p < 0.0001), miR-3687 (8.7-fold; p = 0.0006) and miR-4417 (5.0-fold; p = 0.0005) were most upregulated. While KLK3, miR-21 and miR-141 expression levels in androgen-treated VCaP and LNCaP cells were increased, the expression levels of miR-3687 and miR-4417 were reduced. None of the miRNAs were androgen-regulated in the AR-negative PC3 cell line. Overexpression of miR-3687 reduced cell migration and cell invasion, whilst miR-3195 enhanced cell migration.
Conclusion: We have identified several novel deregulated miRNAs in CRPC tissue, including two microRNAs that are potentially involved in tumor invasion. Our data support the hypothesized involvement of miRNAs in PCa tumorigenesis and progression to CRPC. The applicability of these miRNAs as novel biomarkers for CRPC remains to be further investigated.
Keywords: Androgens; Castration-resistance; MicroRNA; Prostate cancer.
© 2021. The Author(s).
Conflict of interest statement
The authors declare no conflict of interest.
Figures

Similar articles
-
Functional roles and potential clinical application of miRNA-345-5p in prostate cancer.Prostate. 2018 Sep;78(12):927-937. doi: 10.1002/pros.23650. Epub 2018 May 10. Prostate. 2018. PMID: 29748958
-
Investigating miR-6880-5p in extracellular vesicle from plasma as a prognostic biomarker in endocrine therapy-treated castration-resistant prostate cancer.BMC Cancer. 2024 Jul 29;24(1):909. doi: 10.1186/s12885-024-12460-x. BMC Cancer. 2024. PMID: 39075471 Free PMC article.
-
Regulation of NCAPG by miR-99a-3p (passenger strand) inhibits cancer cell aggressiveness and is involved in CRPC.Cancer Med. 2018 May;7(5):1988-2002. doi: 10.1002/cam4.1455. Epub 2018 Apr 2. Cancer Med. 2018. PMID: 29608247 Free PMC article.
-
Role of MicroRNA-21 in Prostate Cancer Progression and Metastasis: Molecular Mechanisms to Therapeutic Targets.Ann Surg Oncol. 2024 Jul;31(7):4795-4808. doi: 10.1245/s10434-024-15453-z. Epub 2024 May 17. Ann Surg Oncol. 2024. PMID: 38758485 Review.
-
MicroRNA Regulation of Androgen Receptor in Castration-Resistant Prostate Cancer: Premises, Promises, and Potentials.Curr Mol Pharmacol. 2021 Oct 25;14(4):559-569. doi: 10.2174/1874467213666201223121850. Curr Mol Pharmacol. 2021. PMID: 33357209 Review.
Cited by
-
Upregulation of microRNA-3687 promotes gestational diabetes mellitus by inhibiting follistatin-like 3.J Perinat Med. 2025 Apr 14;53(5):604-614. doi: 10.1515/jpm-2024-0509. Print 2025 Jun 26. J Perinat Med. 2025. PMID: 40219801
-
miR-3195 suppresses the malignant progression of osteosarcoma cells via targeting SOX4.J Orthop Surg Res. 2023 Oct 30;18(1):809. doi: 10.1186/s13018-023-04321-3. J Orthop Surg Res. 2023. PMID: 37904207 Free PMC article.
-
Identification of the Regulatory Targets of miR-3687 and miR-4417 in Prostate Cancer Cells Using a Proteomics Approach.Int J Mol Sci. 2022 Sep 12;23(18):10565. doi: 10.3390/ijms231810565. Int J Mol Sci. 2022. PMID: 36142477 Free PMC article.
-
The Role of Androgen Receptor and microRNA Interactions in Androgen-Dependent Diseases.Int J Mol Sci. 2022 Jan 28;23(3):1553. doi: 10.3390/ijms23031553. Int J Mol Sci. 2022. PMID: 35163477 Free PMC article. Review.
-
Microphysiological systems as models for immunologically 'cold' tumors.Front Cell Dev Biol. 2024 Apr 22;12:1389012. doi: 10.3389/fcell.2024.1389012. eCollection 2024. Front Cell Dev Biol. 2024. PMID: 38711620 Free PMC article. Review.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

