Retrospective whole-genome comparison of Salmonella enterica serovar Enteritidis from foodborne outbreaks in Southern Brazil
- PMID: 33990934
- PMCID: PMC8324652
- DOI: 10.1007/s42770-021-00508-0
Retrospective whole-genome comparison of Salmonella enterica serovar Enteritidis from foodborne outbreaks in Southern Brazil
Abstract
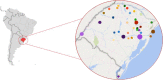
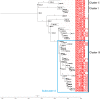
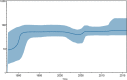
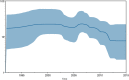
Salmonella enterica serovar Enteritidis is frequently isolated from animal-source foods associated with human salmonellosis outbreaks. This serovar was spread to animal (mainly poultry) farms worldwide in the 1980s, and it is still detected in foods produced in many countries, including Brazil. The present study reports a retrospective genome-wide comparison of S. Enteritidis from foodborne outbreaks in Southern Brazil in the last two decades. Fifty-two S. Enteritidis isolates were obtained from foodborne outbreaks occurring in different cities of the Brazilian southernmost State, Rio Grande do Sul (RS), from 2003 to 2015. Whole-genome sequences (WGS) from these isolates were obtained and comparatively analyzed with 65 additional genomes from NCBI. Phylogenetic and Bayesian analyses were performed to study temporal evolution. Genes related to antibiotic resistance and virulence were also evaluated. The results demonstrated that all S. Enteritidis isolates from Southern Brazil clustered in the global epidemic clade disseminated worldwide originally in the 1980s. Temporal analysis demonstrated that all Brazilian isolates had a tMRCA (time to most recent common ancestor) in 1986 with an effective population size (Ne) increase soon after until 1992, then becoming constant up to now. In Southern Brazil, there was a significant decrease in the spreading of S. Enteritidis in the last decade. In addition, three antibiotic resistance genes were detected in all isolates: aac(6')-Iaa, mdfA, and tet(34). These results demonstrate the high frequency of one only specific S. Enteritidis lineage (global epidemic clade) in foodborne outbreaks from Southern Brazil in the last two decades.
Keywords: Brazil; Foodborne outbreaks; Salmonella enterica.
© 2021. Sociedade Brasileira de Microbiologia.
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- World Organisation for Animal Health (2019) Prevention, detection and control of Salmonella in poultry. https://www.oie.int/index.php?id=169&L=0&htmfile=chapitre_prevent_salmon.... Accessed 07 Feb 2020
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

