Identification and coregulation pattern analysis of long noncoding RNAs following subacute spinal cord injury
- PMID: 33991009
- PMCID: PMC9291281
- DOI: 10.1002/jor.25101
Identification and coregulation pattern analysis of long noncoding RNAs following subacute spinal cord injury
Abstract
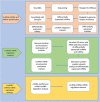
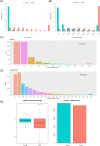
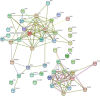
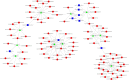
Long noncoding RNAs (lncRNAs) have been demonstrated to play critical regulatory roles in posttranscriptional and transcriptional regulation in eukaryotic cells. However, the characteristics of many lncRNAs, particularly their expression patterns in the lesion epicenter of spinal tissues following subacute spinal cord injury (SCI), remain unclear. In this study, we determined the expression profiles of lncRNAs in the lesion epicenter of spinal tissues after traumatic SCI and predicted latent regulatory networks. Standard Allen's drop surgery was conducted on mice, and hematoxylin and eosin staining was used to observe the damaged area. High-throughput sequencing was performed to identify the differential expression profiles of lncRNAs. Quantitative real-time polymerase chain reaction was conducted to evaluate the quality of the sequencing results. Bioinformatics analyses, including Gene Ontology and Kyoto Encyclopedia of Genes and Genomes pathway analysis, coexpression analysis, and protein-protein interaction analysis, were performed. Targeted binding of lncRNA-miRNA-mRNA was predicted by TargetScan and miRanda. A total of 230 differentially expressed lncRNAs were identified and preliminarily verified, and some potential regulatory networks were constructed. These findings improve our understanding of the mechanisms underlying subacute SCI; differentially expressed lncRNAs are closely involved in pathophysiological processes by regulating multiple pathways. Further studies are essential for revealing the exact mechanism underlying competing endogenous RNA pathways in vivo and in vitro.
Keywords: bioinformatics; long noncoding RNA; regulatory network; subacute spinal cord injury.
© 2021 Journal of Orthopaedic Research® published by Wiley Periodicals LLC on behalf of Orthopaedic Research Society.
Conflict of interest statement
The authors declare that there are no conflict of interests.
Figures





Similar articles
-
Identification of noncoding RNA expression profiles and regulatory interaction networks following traumatic spinal cord injury by sequence analysis.Aging (Albany NY). 2019 Apr 18;11(8):2352-2368. doi: 10.18632/aging.101919. Aging (Albany NY). 2019. PMID: 30998503 Free PMC article.
-
Genome-wide analysis of acute traumatic spinal cord injury-related RNA expression profiles and uncovering of a regulatory axis in spinal fibrotic scars.Cell Prolif. 2021 Jan;54(1):e12951. doi: 10.1111/cpr.12951. Epub 2020 Nov 5. Cell Prolif. 2021. PMID: 33150698 Free PMC article.
-
Expression Profiles of Long Noncoding RNAs and Messenger RNAs in a Rat Model of Spinal Cord Injury.Comput Math Methods Med. 2023 Jan 19;2023:6033020. doi: 10.1155/2023/6033020. eCollection 2023. Comput Math Methods Med. 2023. PMID: 36714328 Free PMC article.
-
Microarray and Bioinformatics Analysis of Differential Gene and lncRNA Expression during Erythropoietin Treatment of Acute Spinal Cord Injury in Rats.Comput Math Methods Med. 2022 Sep 2;2022:4121910. doi: 10.1155/2022/4121910. eCollection 2022. Comput Math Methods Med. 2022. Retraction in: Comput Math Methods Med. 2023 Dec 6;2023:9843413. doi: 10.1155/2023/9843413. PMID: 36092786 Free PMC article. Retracted. Review.
-
Research progress of microRNA in spinal cord injury.Sheng Li Xue Bao. 2024 Jun 25;76(3):394-406. Sheng Li Xue Bao. 2024. PMID: 38939934 Review.
Cited by
-
Lentivirus-mediated overexpression of netrin-1/DCC co-expression promotes axonal regeneration and functional recovery in spinal cord injury via the inhibition of the NgR1-RhoA-ROCK signaling pathway.Transl Neurosci. 2025 Mar 10;16(1):20250365. doi: 10.1515/tnsci-2025-0365. eCollection 2025 Jan 1. Transl Neurosci. 2025. PMID: 40092657 Free PMC article.
-
Identification and coregulation pattern analysis of long noncoding RNAs in the mouse brain after Angiostrongylus cantonensis infection.Parasit Vectors. 2024 May 7;17(1):205. doi: 10.1186/s13071-024-06278-6. Parasit Vectors. 2024. PMID: 38715092 Free PMC article.
References
-
- Harvey LA, Dunlop SA, Churilov L, Galea MP. Spinal Cord Injury Physical Activity (SCIPA) Hands On Trial Collaborators . Early intensive hand rehabilitation is not more effective than usual care plus one‐to‐one hand therapy in people with sub‐acute spinal cord injury ('Hands On'): a randomised trial. J Physiother. 2017;63(4):197‐204. - PubMed
-
- Hara M, Kobayakawa K, Ohkawa Y, et al. Interaction of reactive astrocytes with type I collagen induces astrocytic scar formation through the integrin‐N‐cadherin pathway after spinal cord injury. Nat Med. 2017;23(7):818‐828. - PubMed
-
- Holmes D. Spinal‐cord injury: spurring regrowth. Nature. 2017;552(7684):S49. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

