Identification of a glycolysis-related gene signature associated with clinical outcome for patients with lung squamous cell carcinoma
- PMID: 33991070
- PMCID: PMC8209576
- DOI: 10.1002/cam4.3945
Identification of a glycolysis-related gene signature associated with clinical outcome for patients with lung squamous cell carcinoma
Abstract
Background: Lung squamous cell carcinoma (LUSC), one of the main types of lung cancer, has caused a huge social burden. There has been no significant progress in its therapy in recent years, Resulting in a poor prognosis. This study aims to develop a glycolysis-related gene signature to predict patients' survival with LUSC and explore new therapeutic targets.
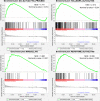
Methods: We obtained the mRNA expression and clinical information of 550 patients with LUSC from the Cancer Genome Atlas (TCGA) database. Glycolysis genes were identified by Gene Set Enrichment Analysis (GSEA). The glycolysis-related gene signature was established using the Cox regression analysis.
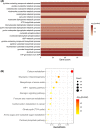
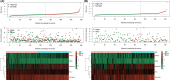
Results: We developed five glycolysis-related genes signature (HKDC1, AGL, ALDH7A1, SLC16A3, and MIOX) to calculate each patient's risk score. According to the risk score, patients were divided into high- and low-risk groups and exhibited significant differences in overall survival (OS) between the two groups. The ROC curves showed that the AUC was 0.707 for the training cohort and 0.651 for the validation cohort. Additionally, the risk score was confirmed as an independent risk factor for LUSC patients by Cox regression analysis.
Conclusion: We built a gene signature to clarify the connection between glycolysis and LUSC. This model performs well in evaluating patients' survival with LUSC and provides new biomarkers for targeted therapy.
Keywords: gene signature; glycolysis; lung squamous cell carcinoma; survival.
© 2021 The Authors. Cancer Medicine published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors have no conflict of interest to declare.
Figures

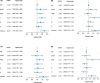
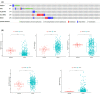
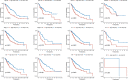


References
-
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7‐30. - PubMed
-
- Qi L, Gao C, Feng F, et al. MicroRNAs associated with lung squamous cell carcinoma: new prognostic biomarkers and therapeutic targets. J Cell Biochem. 2019;120(11):18956‐18966. - PubMed
-
- Xu Y, Li J, Wang P, Zhang Z, Wang X. LncRNA HULC promotes lung squamous cell carcinoma by regulating PTPRO via NF‐kappaB. J Cell Biochem. 2019;120(12):19415‐19421. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

