Protein-based lateral flow assays for COVID-19 detection
- PMID: 33991088
- PMCID: PMC8194834
- DOI: 10.1093/protein/gzab010
Protein-based lateral flow assays for COVID-19 detection
Abstract
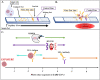
To combat the enduring and dangerous spread of COVID-19, many innovations to rapid diagnostics have been developed based on proteinprotein interactions of the SARS-CoV-2 spike and nucleocapsid proteins to increase testing accessibility. These antigen tests have most prominently been developed using the lateral flow assay (LFA) test platform which has the benefit of administration at point-of-care, delivering quick results, lower cost, and does not require skilled personnel. However, they have gained criticism for an inferior sensitivity. In the last year, much attention has been given to creating a rapid LFA test for detection of COVID-19 antigens that can address its high limit of detection while retaining the advantages of rapid antibodyantigen interaction. In this review, a summary of these proteinprotein interactions as well as the challenges, benefits, and recent improvements to protein based LFA for detection of COVID-19 are discussed.
Keywords: antibody test; antigen test; covid-19; diagnostics; lateral flow assay.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

References
-
- Abudayyeh, O.O., Gootenberg, J.S., Konermann, S.et al. (2016) Science, 353, aaf5573-1 to aaf5573-9.
-
- Alexandersen, S., Chamings, A., Bhatta, T.R. (2020) medRxiv.
-
- Ariffin, N., Yusof, N.A., Abdullah, J., Rahman, S.F.A., Raston, N.H.A., Kusnin, N., Suraiya, S. (2020) J. Sensors, 2020, 1365983.
-
- Arnaout, R., Lee, R.A., Lee, G.R., Callahan, C., Yen, C.F., Smith, K.P., Arora, R., Kirby, J.E. (2020) bioRxiv.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

