Genetic potential for residual feed intake and diet fed during early- to mid-gestation influences post-natal DNA methylation of imprinted genes in muscle and liver tissues in beef cattle
- PMID: 33991189
- PMCID: PMC8160533
- DOI: 10.1093/jas/skab140
Genetic potential for residual feed intake and diet fed during early- to mid-gestation influences post-natal DNA methylation of imprinted genes in muscle and liver tissues in beef cattle
Abstract
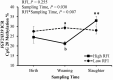
Discovery of epigenetic modifications associated with feed efficiency or other economically important traits would increase our understanding of the molecular mechanisms underlying these traits. In combination with known genetic markers, this would provide opportunity to improve genomic selection accuracy in cattle breeding programs. It would also allow cattle to be managed to improve favorable gene expression. The objective of this study was to identify variation in DNA methylation between beef cattle of differential pre-natal nutrition and divergent genetic potential for residual feed intake (RFI). Purebred Angus offspring with the genetic potential for either high (HRFI) or low (LRFI) RFI were prenatally exposed to either a restricted maternal diet of 0.5 kg/d average daily gain (ADG) or a moderate maternal diet of 0.7 kg/d ADG from 30 to 150 d of gestation. We performed DNA methylation analysis of differentially methylated regions (DMR) of imprinted genes (Insulin-like growth factor 2 (IGF2) DMR2, IGF2/H19 imprinting control region (ICR) and IGF2 receptor (IGF2R) DMR2) using post-natal samples of longissimus dorsi (LD) muscle taken from male and female calves at birth and weaning, and of LD muscle, semimembranosus (SM) muscle, and liver samples collected from steers at slaughter (17 months of age). Interestingly, for all three DMR investigated in liver, LRFI steers had higher levels of methylation than HRFI steers. In LD muscle, IGF2/H19 ICR methylation differences for heifers at birth were due to pre-natal diet, while for steers at birth they were mostly the result of genetic potential for RFI with LRFI steers again having higher levels of methylation than HRFI steers. While results from repeated measures analysis of DNA methylation in steers grouped by RFI revealed few differences, in steers grouped by diet, we found higher methylation levels of IGF2 DMR2 and IGF2R DMR2 in LD muscle of restricted diet steers at weaning and slaughter than at birth, as well as increased methylation in LD muscle of restricted diet steers compared with moderate diet steers at weaning and/or slaughter. Our results suggest that differential pre-natal nutrition, and divergent genetic potential for RFI, induces tissue- and sex-specific alterations in post-natal IGF2 and IGF2R methylation patterns and that these patterns can vary with age in Angus beef cattle.
Keywords: beef cattle; epigenetics; imprinted genes; methylation; pre-natal diet; residual feed intake.
© Her Majesty the Queen in Right of Canada, as represented by the Minister of the Department of Agriculture and Agri-Food, Government of Canada, 2021.
Figures


References
-
- Alberta Agriculture and Forestry, Government of Alberta . 2019. 2014–2018 Economic, Productive and Financial Performance of Alberta Cow/Calf Operations. https://open.alberta.ca/publications/agriprofit-economic-production-and-.... (Accessed 27 November 2020).
-
- Abo-Ismail, M. K., Lansink N., Akanno E., Karisa B. K., Crowley J. J., Moore S. S., Bork E., Stothard P., Basarab J. A., and Plastow G. S.. . 2018. Development and validation of a small SNP panel for feed efficiency in beef cattle. J. Anim. Sci. 96:375–397. doi: 10.1093/jas/sky020 - DOI - PMC - PubMed
-
- Abo-Ismail, M. K., Vander Voort G., Squires J. J., Swanson K. C., Mandell I. B., Liao X., Stothard P., Moore S., Plastow G., and Miller S. P.. . 2014. Single nucleotide polymorphisms for feed efficiency and performance in crossbred beef cattle. BMC Genet. 15:14. doi: 10.1186/1471-2156-15-14 - DOI - PMC - PubMed
-
- Arthur, P. F., Renand G., and Krauss D.. . 2001. Genetic and phenotypic relationships among different measures of growth and feed efficiency in young Charolais bulls. Livest. Prod. Sci. 68(2):131–139. doi: 10.1016/S0301-6226(00)00243-8 - DOI
-
- Basarab, J. A., Price M. A., Aalhus J. L., Okine E. K., Snelling W. M., and Lyle K. L.. . 2003. Residual feed intake and body composition in young growing cattle. Can. J. Anim. Sci. 83(2):189–204. doi: 10.4141/a02-065 - DOI
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

