The nucleotide binding affinities of two critical conformations of Escherichia coli ATP synthase
- PMID: 33991499
- PMCID: PMC8278868
- DOI: 10.1016/j.abb.2021.108899
The nucleotide binding affinities of two critical conformations of Escherichia coli ATP synthase
Abstract
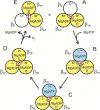
ATP synthase is essential in aerobic energy metabolism, and the rotary catalytic mechanism is one of the core concepts to understand the energetic functions of ATP synthase. Disulfide bonds formed by oxidizing a pair of cysteine mutations halted the rotation of the γ subunit in two critical conformations, the ATP-waiting dwell (αE284C/γQ274C) and the catalytic dwell (αE284C/γL276C). Tryptophan fluorescence was used to measure the nucleotide binding affinities for MgATP, MgADP and MgADP-AlF4 (a transition state analog) to wild-type and mutant F1 under reducing and oxidizing conditions. In the reduced state, αE284C/γL276C F1 showed a wild-type-like nucleotide binding pattern; after oxidation to lock the enzyme in the catalytic dwell state, the nucleotide binding parameters remained unchanged. In contrast, αE284C/γQ274C F1 showed significant differences in the affinities of the oxidized versus the reduced state. Locking the enzyme in the ATP-waiting dwell reduced nucleotide binding affinities of all three catalytic sites. Most importantly, the affinity of the low affinity site was reduced to such an extent that it could no longer be detected in the binding assay (Kd > 5 mM). The results of the present study allow to present a model for the catalytic mechanism of ATP synthase under consideration of the nucleotide affinity changes during a 360° cycle of the rotor.
Keywords: ATP synthase; Catalytic mechanism; Disulfide crosslink; Nucleotide binding; Rotational catalysis; Tryptophan fluorescence.
Copyright © 2021 Elsevier Inc. All rights reserved.
Conflict of interest statement
Conflict of interest
The authors declare that there is no conflict of interest.
Figures



References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

