Epitope profiling reveals binding signatures of SARS-CoV-2 immune response in natural infection and cross-reactivity with endemic human CoVs
- PMID: 33991511
- PMCID: PMC8121454
- DOI: 10.1016/j.celrep.2021.109164
Epitope profiling reveals binding signatures of SARS-CoV-2 immune response in natural infection and cross-reactivity with endemic human CoVs
Abstract
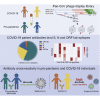
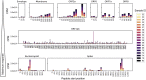
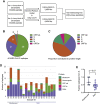
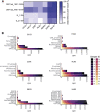
A major goal of current severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) vaccine efforts is to elicit antibody responses that confer protection. Mapping the epitope targets of the SARS-CoV-2 antibody response is critical for vaccine design, diagnostics, and development of therapeutics. Here, we develop a pan-coronavirus phage display library to map antibody binding sites at high resolution within the complete viral proteomes of all known human-infecting coronaviruses in patients with mild or moderate/severe coronavirus disease 2019 (COVID-19). We find that the majority of immune responses to SARS-CoV-2 are targeted to the spike protein, nucleocapsid, and ORF1ab and include sites of mutation in current variants of concern. Some epitopes are identified in the majority of samples, while others are rare, and we find variation in the number of epitopes targeted between individuals. We find low levels of SARS-CoV-2 cross-reactivity in individuals with no exposure to the virus and significant cross-reactivity with endemic human coronaviruses (CoVs) in convalescent sera from patients with COVID-19.
Keywords: COVID-19; SARS-CoV-2; cross-reactivity; epitopes; phage-display; serology; variants.
Copyright © 2021 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests The authors declare no competing interests.
Figures



References
-
- Arvin A.M., Fink K., Schmid M.A., Cathcart A., Spreafico R., Havenar-Daughton C., Lanzavecchia A., Corti D., Virgin H.W. A perspective on potential antibody-dependent enhancement of SARS-CoV-2. Nature. 2020;584:353–363. - PubMed
-
- Chan K.H., Cheng V.C.C., Woo P.C.Y., Lau S.K.P., Poon L.L.M., Guan Y., Seto W.H., Yuen K.Y., Peiris J.S.M. Serological responses in patients with severe acute respiratory syndrome coronavirus infection and cross-reactivity with human coronaviruses 229E, OC43, and NL63. Clin. Diagn. Lab. Immunol. 2005;12:1317–1321. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases
Miscellaneous

