Effect of mismatch repair on the mutational footprint of the bacterial SOS mutator activity
- PMID: 33991871
- PMCID: PMC8206030
- DOI: 10.1016/j.dnarep.2021.103130
Effect of mismatch repair on the mutational footprint of the bacterial SOS mutator activity
Abstract
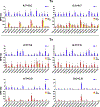
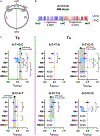
The bacterial SOS response to DNA damage induces an error-prone repair program that is mutagenic. In Escherichia coli, SOS-induced mutations are caused by the translesion synthesis (TLS) activity of two error-prone polymerases (EPPs), Pol IV and Pol V. The mutational footprint of the EPPs is confounded by both DNA damage and repair, as mutations are targeted to DNA lesions via TLS and corrected by the mismatch repair (MMR) system. To remove these factors and assess untargeted EPP mutations genome-wide, we constructed spontaneous SOS mutator strains deficient in MMR, then analyzed their mutational footprints by mutation accumulation and whole genome sequencing. Our analysis reveals new features of untargeted SOS-mutagenesis, showing how MMR alters its spectrum, sequence specificity, and strand-bias. Our data support a model where the EPPs prefer to act on the lagging strand of the replication fork, producing base pair mismatches that are differentially repaired by MMR depending on the type of mismatch.
Keywords: DNA polymerase IV; DNA polymerase V; Error-prone polymerases; Mismatch repair; Mutation accumulation whole genome sequencing; SOS response.
Copyright © 2021 Elsevier B.V. All rights reserved.
Figures



References
-
- Friedberg EC, Walker GC, Siede W, DNA repair and mutagenesis, ASM Press, Washington, D.C., 1995.
-
- Tang M, Pham P, Shen X, Taylor JS, O’Donnell M, Woodgate R, Goodman MF, Roles of E. coli DNA polymerases IV and V in lesion-targeted and untargeted SOS mutagenesis, Nature, 404 (2000) 1014–1018. - PubMed
-
- Kobayashi S, Valentine MR, Pham P, O’Donnell M, Goodman MF, Fidelity of Escherichia coli DNA polymerase IV. Preferential generation of small deletion mutations by dNTP-stabilized misalignment, J Biol Chem, 277 (2002) 34198–34207. - PubMed
-
- Jarosz DF, Beuning PJ, Cohen SE, Walker GC, Y-family DNA polymerases in Escherichia coli, Trends in microbiology, 15 (2007) 70–77. - PubMed
Publication types
MeSH terms
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

