Cauliflower mosaic virus protein P6-TAV plays a major role in alteration of aphid vector feeding behaviour but not performance on infected Arabidopsis
- PMID: 33993609
- PMCID: PMC8295513
- DOI: 10.1111/mpp.13069
Cauliflower mosaic virus protein P6-TAV plays a major role in alteration of aphid vector feeding behaviour but not performance on infected Arabidopsis
Abstract
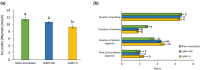
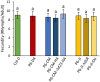
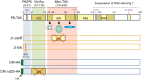
Emerging evidence suggests that viral infection modifies host plant traits that in turn alter behaviour and performance of vectors colonizing the plants in a way conducive for transmission of both nonpersistent and persistent viruses. Similar evidence for semipersistent viruses like cauliflower mosaic virus (CaMV) is scarce. Here we compared the effects of Arabidopsis infection with mild (CM) and severe (JI) CaMV isolates on the feeding behaviour (recorded by the electrical penetration graph technique) and fecundity of the aphid vector Myzus persicae. Compared to mock-inoculated plants, feeding behaviour was altered similarly on CM- and JI-infected plants, but only aphids on JI-infected plants had reduced fecundity. To evaluate the role of the multifunctional CaMV protein P6-TAV, aphid feeding behaviour and fecundity were tested on transgenic Arabidopsis plants expressing wild-type (wt) and mutant versions of P6-TAV. In contrast to viral infection, aphid fecundity was unchanged on all transgenic lines, suggesting that other viral factors compromise fecundity. Aphid feeding behaviour was modified on wt P6-CM-, but not on wt P6-JI-expressing plants. Analysis of plants expressing P6 mutants identified N-terminal P6 domains contributing to modification of feeding behaviour. Taken together, we show that CaMV infection can modify both aphid fecundity and feeding behaviour and that P6 is only involved in the latter.
Keywords: aphid vector; plant virus; vector behaviour; vector modification; vector transmission; viral factors.
© 2021 The Authors. Molecular Plant Pathology published by British Society for Plant Pathology and John Wiley & Sons Ltd.
Figures



Similar articles
-
Plant Viruses Can Alter Aphid-Triggered Calcium Elevations in Infected Leaves.Cells. 2021 Dec 14;10(12):3534. doi: 10.3390/cells10123534. Cells. 2021. PMID: 34944040 Free PMC article.
-
The cauliflower mosaic virus transmission helper protein P2 modifies directly the probing behavior of the aphid vector Myzus persicae to facilitate transmission.PLoS Pathog. 2023 Feb 6;19(2):e1011161. doi: 10.1371/journal.ppat.1011161. eCollection 2023 Feb. PLoS Pathog. 2023. PMID: 36745680 Free PMC article.
-
Comparative Plant Transcriptome Profiling of Arabidopsis thaliana Col-0 and Camelina sativa var. Celine Infested with Myzus persicae Aphids Acquiring Circulative and Noncirculative Viruses Reveals Virus- and Plant-Specific Alterations Relevant to Aphid Feeding Behavior and Transmission.Microbiol Spectr. 2022 Aug 31;10(4):e0013622. doi: 10.1128/spectrum.00136-22. Epub 2022 Jul 20. Microbiol Spectr. 2022. PMID: 35856906 Free PMC article.
-
A model for intracellular movement of Cauliflower mosaic virus: the concept of the mobile virion factory.J Exp Bot. 2016 Mar;67(7):2039-48. doi: 10.1093/jxb/erv520. Epub 2015 Dec 18. J Exp Bot. 2016. PMID: 26687180 Review.
-
Modelling and manipulation of aphid-mediated spread of non-persistently transmitted viruses.Virus Res. 2020 Feb;277:197845. doi: 10.1016/j.virusres.2019.197845. Epub 2019 Dec 23. Virus Res. 2020. PMID: 31874210 Free PMC article. Review.
Cited by
-
The Turnip Yellows Virus Capsid Protein Promotes Access of Its Main Aphid Vector Myzus persicae to Phloem Tissues.Plant Cell Environ. 2025 Mar;48(3):2434-2444. doi: 10.1111/pce.15303. Epub 2024 Dec 2. Plant Cell Environ. 2025. PMID: 39623721 Free PMC article.
-
Plant Viruses Can Alter Aphid-Triggered Calcium Elevations in Infected Leaves.Cells. 2021 Dec 14;10(12):3534. doi: 10.3390/cells10123534. Cells. 2021. PMID: 34944040 Free PMC article.
-
The cauliflower mosaic virus transmission helper protein P2 modifies directly the probing behavior of the aphid vector Myzus persicae to facilitate transmission.PLoS Pathog. 2023 Feb 6;19(2):e1011161. doi: 10.1371/journal.ppat.1011161. eCollection 2023 Feb. PLoS Pathog. 2023. PMID: 36745680 Free PMC article.
-
Comparative Plant Transcriptome Profiling of Arabidopsis thaliana Col-0 and Camelina sativa var. Celine Infested with Myzus persicae Aphids Acquiring Circulative and Noncirculative Viruses Reveals Virus- and Plant-Specific Alterations Relevant to Aphid Feeding Behavior and Transmission.Microbiol Spectr. 2022 Aug 31;10(4):e0013622. doi: 10.1128/spectrum.00136-22. Epub 2022 Jul 20. Microbiol Spectr. 2022. PMID: 35856906 Free PMC article.
References
-
- Blanc, S. , Cerutti, M. , Usmany, M. , Vlak, J.M. & Hull, R. (1993) Biological activity of cauliflower mosaic virus aphid transmission factor expressed in a heterologous system. Virology, 192, 643–650. - PubMed
-
- Bouchery, Y. , Givord, L. & Monestiez, P. (1990) Comparison of short‐ and long‐feed transmission of the cauliflower mosaic virus Cabb‐S strain and S delta II hybrid by two species of aphid: Myzus persicae (Sulzer) and Brevicoryne brassicae (L.). Research in Virology, 141, 677–683. - PubMed
-
- Cecchini, E. , Al‐Kaff, N.S. , Bannister, A. , Giannakou, M.E. , McCallum, D.G. , Maule, A.J. et al. (1998) Pathogenic interactions between variants of cauliflower mosaic virus and Arabidopsis thaliana . Journal of Experimental Botany, 49, 731–737.
-
- Champagne, J. , Benhamou, N. & Leclerc, D. (2004) Localization of the N‐terminal domain of cauliflower mosaic virus coat protein precursor. Virology, 324, 257–262. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

