SARS-CoV-2 RNA screening in routine pathology specimens
- PMID: 33993637
- PMCID: PMC8209898
- DOI: 10.1111/1751-7915.13828
SARS-CoV-2 RNA screening in routine pathology specimens
Abstract
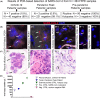
Virus detection methods are important to cope with the SARS-CoV-2 pandemics. Apart from the lung, SARS-CoV-2 was detected in multiple organs in severe cases. Less is known on organ tropism in patients developing mild or no symptoms, and some of such patients might be missed in symptom-indicated swab testing. Here, we tested and validated several approaches and selected the most reliable RT-PCR protocol for the detection of SARS-CoV-2 RNA in patients' routine diagnostic formalin-fixed and paraffin-embedded (FFPE) specimens available in pathology, to assess (i) organ tropism in samples from COVID-19-positive patients, (ii) unrecognized cases in selected tissues from negative or not-tested patients during a pandemic peak, and (iii) retrospectively, pre-pandemic lung samples. We identified SARS-CoV-2 RNA in seven samples from confirmed COVID-19 patients, in two gastric biopsies, one small bowel and one colon resection, one lung biopsy, one pleural resection and one pleural effusion specimen, while all other specimens were negative. In the pandemic peak cohort, we identified one previously unrecognized COVID-19 case in tonsillectomy samples. All pre-pandemic lung samples were negative. In conclusion, SARS-CoV-2 RNA detection in FFPE pathology specimens can potentially improve surveillance of COVID-19, allow retrospective studies, and advance our understanding of SARS-CoV-2 organ tropism and effects.
© 2021 The Authors. Microbial Biotechnology published by John Wiley & Sons Ltd and Society for Applied Microbiology.
Conflict of interest statement
The authors declare that there is nothing to disclose.
Figures


References
Publication types
MeSH terms
Substances
Grants and funding
- ZMVI1-2520COR201/Bundesministerium für Gesundheit
- BO3755/13-1 - Project-ID 454024652/Deutsche Forschungsgemeinschaft
- BO3755/9-1 - Project-ID 432698239/Deutsche Forschungsgemeinschaft
- SFB/TRR219 - Project-ID 322900939/Deutsche Forschungsgemeinschaft
- SFB/TRR57- Project ID 36842431/Deutsche Forschungsgemeinschaft
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

