An insight into the inhibitory mechanism of phytochemicals and FDA-approved drugs on the ACE2-Spike complex of SARS-CoV-2 using computational methods
- PMID: 33994655
- PMCID: PMC8106519
- DOI: 10.1007/s11696-021-01680-1
An insight into the inhibitory mechanism of phytochemicals and FDA-approved drugs on the ACE2-Spike complex of SARS-CoV-2 using computational methods
Abstract
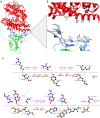
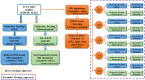
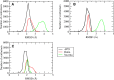
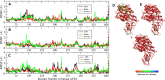
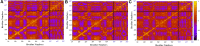
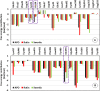
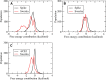
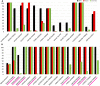
The S-glycoprotein (Spike) of the SARS-CoV-2 forms a complex with the human transmembrane protein angiotensin-converting enzyme 2 (ACE2) during infection. It forms the first line of contact with the human cell. The FDA-approved drugs and phytochemicals from Indian medicinal plants were explored. Molecular docking and simulations of these molecules targeting the ACE2-Spike complex were performed. Rutin DAB10 and Swertiapuniside were obtained as the top-scored drugs as per the docking protocol. The MD simulations of ligand-free, Rutin DAB10-bound, and Swertiapuniside-bound ACE2-Spike complex revealed abrogation of the hydrogen bonding network between the two proteins. The principal component and dynamic cross-correlation analysis pointed out conformational changes in both the proteins unique to the ligand-bound systems. The interface residues, His34, and Lys353 from ACE2 and Arg403, and Tyr495 from the Spike protein formed significant strong interactions with the ligand molecules, inferring the inhibition of ACE2-Spike complex. Few novel interactions specific to Rutin-DAB10 and Swertiapuniside were also identified. The conformational flexibility of the drug-binding pocket was captured using the RMSD-based clustering of the ligand-free simulations. Ensemble docking was performed wherein the FDA-approved database and phytochemical dataset were docked on each of the cluster representatives of the ACE2-Spike. The phytochemicals identified belonged to Withania somnifera, Swertia chirayita, Tinospora cordifolia and Rutin DAB10, fulvestrant, elbasvir from FDA.
Supplementary information: The online version contains supplementary material available at 10.1007/s11696-021-01680-1.
Keywords: ACE2; FDA; Molecular dynamics; Phytochemicals; Spike protein.
© Institute of Chemistry, Slovak Academy of Sciences 2021.
Conflict of interest statement
Conflict of interestThe authors declare no conflict of interest.
Figures






References
-
- Alam N, Hossain M, Khalil MI, et al. Recent advances in elucidating the biological properties of Withania somnifera and its potential role in health benefits. Phytochem Rev. 2012;11:97–112. doi: 10.1007/s11101-011-9221-5. - DOI
-
- Case DA, Betz RM, Cerutti DS, Cheatham TE, III, Darden TA, Duke RE, Giese TJ, Gohlke H, Goetz AW, Homeyer N, Izadi S, Janowski P, Kaus J, Kovalenko A, Lee TS, LeGrand S, Li P, Lin C, Luchko T, Luo R, Madej B, Mermelstein D, Merz KM, Monard G, Nguyen H, Nguyen HT, Omelyan I, Onufriev A, Roe DR, Roitberg A, Sagui C, Simmerling CL, Botello-Smith WM, Swails J, Walker RC, Wang J, Wolf RM, Wu X, Xiao L, Kollman PA. AMBER 2016. San Francisco: University of California; 2016.
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous
