Spatial Memory and Gut Microbiota Alterations Are Already Present in Early Adulthood in a Pre-clinical Transgenic Model of Alzheimer's Disease
- PMID: 33994914
- PMCID: PMC8116633
- DOI: 10.3389/fnins.2021.595583
Spatial Memory and Gut Microbiota Alterations Are Already Present in Early Adulthood in a Pre-clinical Transgenic Model of Alzheimer's Disease
Abstract
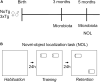
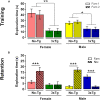
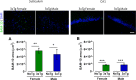
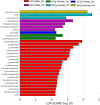
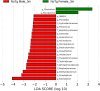
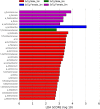
The irreversible and progressive neurodegenerative Alzheimer's disease (AD) is characterized by cognitive decline, extracellular β-amyloid peptide accumulation, and tau neurofibrillary tangles in the cortex and hippocampus. The triple-transgenic (3xTg) mouse model of AD presents memory impairment in several behavioral paradigms and histopathological alterations from 6 to 16 months old. Additionally, it seems that dysbiotic gut microbiota is present in both mouse models and patients of AD at the cognitive symptomatic stage. The present study aimed to assess spatial learning, memory retention, and gut microbiota alterations in an early adult stage of the 3xTg-AD mice as well as to explore its sexual dimorphism. We evaluated motor activity, novel-object localization training, and retention test as well as collected fecal samples to characterize relative abundance, alpha- and beta-diversity, and linear discriminant analysis (LDA) effect size (LEfSe) analysis in gut microbiota in both female and male 3xTg-AD mice, and controls [non-transgenic mice (NoTg)], at 3 and 5 months old. We found spatial memory deficits in female and male 3xTg-AD but no alteration neither during training nor in motor activity. Importantly, already at 3 months old, we observed decreased relative abundances of Actinobacteria and TM7 in 3xTg-AD compared to NoTg mice, while the beta diversity of gut microbiota was different in female and male 3xTg-AD mice in comparison to NoTg. Our results suggest that gut microbiota modifications in 3xTg-AD mice anticipate and thus could be causally related to cognitive decline already at the early adult age of AD. We propose that microbiota alterations may be used as an early and non-invasive diagnostic biomarker of AD.
Keywords: 3xTg-AD; Actinobacteria; TM7; alpha-diversity; beta-diversity; dysbiosis; high-throughput DNA sequencing; novel-object localization.
Copyright © 2021 Bello-Medina, Hernández-Quiroz, Pérez-Morales, González-Franco, Cruz-Pauseno, García-Mena, Díaz-Cintra and Pacheco-López.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures







Similar articles
-
Alterations of Spatial Memory and Gut Microbiota Composition in Alzheimer's Disease Triple-Transgenic Mice at 3, 6, and 9 Months of Age.Am J Alzheimers Dis Other Demen. 2023 Jan-Dec;38:15333175231174193. doi: 10.1177/15333175231174193. Am J Alzheimers Dis Other Demen. 2023. PMID: 37169734 Free PMC article.
-
Alterations in synaptic plasticity coincide with deficits in spatial working memory in presymptomatic 3xTg-AD mice.Neurobiol Learn Mem. 2015 Nov;125:152-162. doi: 10.1016/j.nlm.2015.09.003. Epub 2015 Sep 15. Neurobiol Learn Mem. 2015. PMID: 26385257 Free PMC article.
-
Predicting Neurodegenerative Disease Using Prepathology Gut Microbiota Composition: a Longitudinal Study in Mice Modeling Alzheimer's Disease Pathologies.Microbiol Spectr. 2023 Mar 6;11(2):e0345822. doi: 10.1128/spectrum.03458-22. Online ahead of print. Microbiol Spectr. 2023. PMID: 36877047 Free PMC article.
-
Herbal medicines in Alzheimer's disease and the involvement of gut microbiota.Front Pharmacol. 2024 Jul 16;15:1416502. doi: 10.3389/fphar.2024.1416502. eCollection 2024. Front Pharmacol. 2024. PMID: 39081953 Free PMC article. Review.
-
Anxiety and Alzheimer's disease pathogenesis: focus on 5-HT and CRF systems in 3xTg-AD and TgF344-AD animal models.Front Aging Neurosci. 2023 Nov 10;15:1251075. doi: 10.3389/fnagi.2023.1251075. eCollection 2023. Front Aging Neurosci. 2023. PMID: 38076543 Free PMC article. Review.
Cited by
-
Cognitive Effects of Simulated Galactic Cosmic Radiation Are Mediated by ApoE Status, Sex, and Environment in APP Knock-In Mice.Int J Mol Sci. 2024 Aug 29;25(17):9379. doi: 10.3390/ijms25179379. Int J Mol Sci. 2024. PMID: 39273325 Free PMC article.
-
Chronic-Antibiotics Induced Gut Microbiota Dysbiosis Rescues Memory Impairment and Reduces β-Amyloid Aggregation in a Preclinical Alzheimer's Disease Model.Int J Mol Sci. 2022 Jul 26;23(15):8209. doi: 10.3390/ijms23158209. Int J Mol Sci. 2022. PMID: 35897785 Free PMC article.
-
Microbiota in neuroinflammation and synaptic dysfunction: a focus on Alzheimer's disease.Mol Neurodegener. 2022 Mar 5;17(1):19. doi: 10.1186/s13024-022-00522-2. Mol Neurodegener. 2022. PMID: 35248147 Free PMC article. Review.
-
The Potential Role of Gut Microbiota in Alzheimer's Disease: From Diagnosis to Treatment.Nutrients. 2022 Feb 5;14(3):668. doi: 10.3390/nu14030668. Nutrients. 2022. PMID: 35277027 Free PMC article. Review.
-
Soybean Meal Extract Preserves Memory Ability by Increasing Presynaptic Function and Modulating Gut Microbiota in Rats.Mol Neurobiol. 2022 Mar;59(3):1649-1664. doi: 10.1007/s12035-021-02669-3. Epub 2022 Jan 10. Mol Neurobiol. 2022. PMID: 35001354
References
-
- Andrews S. (2010). FastQC: A Quality Control Tool for High Throughput Sequence Data. Available online at: http://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed July, 2020).
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

