Transductive Transfer Learning for Domain Adaptation in Brain Magnetic Resonance Image Segmentation
- PMID: 33994917
- PMCID: PMC8116893
- DOI: 10.3389/fnins.2021.608808
Transductive Transfer Learning for Domain Adaptation in Brain Magnetic Resonance Image Segmentation
Abstract
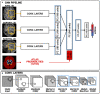
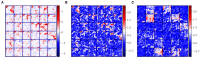
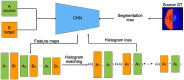
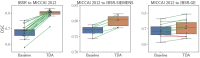
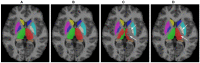
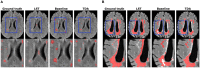
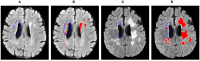
Segmentation of brain images from Magnetic Resonance Images (MRI) is an indispensable step in clinical practice. Morphological changes of sub-cortical brain structures and quantification of brain lesions are considered biomarkers of neurological and neurodegenerative disorders and used for diagnosis, treatment planning, and monitoring disease progression. In recent years, deep learning methods showed an outstanding performance in medical image segmentation. However, these methods suffer from generalisability problem due to inter-centre and inter-scanner variabilities of the MRI images. The main objective of the study is to develop an automated deep learning segmentation approach that is accurate and robust to the variabilities in scanner and acquisition protocols. In this paper, we propose a transductive transfer learning approach for domain adaptation to reduce the domain-shift effect in brain MRI segmentation. The transductive scenario assumes that there are sets of images from two different domains: (1) source-images with manually annotated labels; and (2) target-images without expert annotations. Then, the network is jointly optimised integrating both source and target images into the transductive training process to segment the regions of interest and to minimise the domain-shift effect. We proposed to use a histogram loss in the feature level to carry out the latter optimisation problem. In order to demonstrate the benefit of the proposed approach, the method has been tested in two different brain MRI image segmentation problems using multi-centre and multi-scanner databases for: (1) sub-cortical brain structure segmentation; and (2) white matter hyperintensities segmentation. The experiments showed that the segmentation performance of a pre-trained model could be significantly improved by up to 10%. For the first segmentation problem it was possible to achieve a maximum improvement from 0.680 to 0.799 in average Dice Similarity Coefficient (DSC) metric and for the second problem the average DSC improved from 0.504 to 0.602. Moreover, the improvements after domain adaptation were on par or showed better performance compared to the commonly used traditional unsupervised segmentation methods (FIRST and LST), also achieving faster execution time. Taking this into account, this work presents one more step toward the practical implementation of deep learning algorithms into the clinical routine.
Keywords: brain; deep learning; domain adaptation; magnetic resonance imaging; segmentation; sub-cortical structures; transductive learning; white matter hyperintensities.
Copyright © 2021 Kushibar, Salem, Valverde, Rovira, Salvi, Oliver and Lladó.
Conflict of interest statement
ÀR serves on scientific advisory boards for Novartis, Sanofi-Genzyme, Icometrix, SyntheticMR, and OLEA Medical, and has received speaker honoraria from Bayer, Sanofi-Genzyme, Bracco, Merck-Serono, Teva Pharmaceutical Industries Ltd, Novartis, Roche, and Biogen Idec. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
-
- Bakas S., Reyes M., Jakab A., Bauer S., Rempfler M., Crimi A., et al. . (2018). Identifying the best machine learning algorithms for brain tumor segmentation, progression assessment, and overall survival prediction in the BRATS challenge. arXiv [Preprint]. arXiv:1811.02629.
-
- Campello M., Lekadir K. (2020). “Multi-centre multi-vendor & multi-disease cardiac image segmentation challenge (M&Ms),” in Medical Image Computing and Computer Assisted Intervention. (Lima: ).
LinkOut - more resources
Full Text Sources
Other Literature Sources

