Novel Insight Into the Role of Immune Dysregulation in Amyotrophic Lateral Sclerosis Based on Bioinformatic Analysis
- PMID: 33994932
- PMCID: PMC8119763
- DOI: 10.3389/fnins.2021.657465
Novel Insight Into the Role of Immune Dysregulation in Amyotrophic Lateral Sclerosis Based on Bioinformatic Analysis
Abstract
Amyotrophic lateral sclerosis (ALS) is a fatal neurodegenerative disorder characterized by the progressive degeneration of motor neurons. The causative pathogenic mechanisms in ALS remain unclear, limiting the development of treatment strategies. Neuroinflammation and immune dysregulation were involved in the disease onset and progression of several neurodegenerative disorders, including ALS. In this study, we carried out a bioinformatic analysis using publicly available datasets from Gene Expression Omnibus (GEO) to investigate the role of immune cells and genes alterations in ALS. Single-sample gene set enrichment analysis revealed that the infiltration of multiple types of immune cells, including macrophages, type-1/17 T helper cells, and activated CD4 + /CD8 + T cells, was higher in ALS patients than in controls. Weighted gene correlation network analysis identified immune genes associated with ALS. The Gene Ontology analysis revealed that receptor and cytokine activities were the most highly enriched terms. Pathway analysis showed that these genes were enriched not only in immune-related pathways, such as cytokine-cytokine receptor interaction, but also in PI3K-AKT and MAPK signaling pathways. Nineteen immune-related genes (C3AR1, CCR1, CCR5, CD86, CYBB, FCGR2B, FCGR3A, HCK, ITGB2, PTPRC, TLR1, TLR2, TLR7, TLR8, TYROBP, VCAM1, CD14, CTSS, and FCER1G) were identified as hub genes based on least absolute shrinkage and selection operator analysis. This gene signature could differentiate ALS patients from non-neurological controls (p < 0.001) and predict disease occurrence (AUC = 0.829 in training set; AUC = 0.862 in test set). In conclusion, our study provides potential biomarkers of ALS for disease diagnosis and therapeutic monitoring.
Keywords: LASSO; WGCNA; amyotrophic lateral sclerosis; bioinformatics; immune; ssGSEA.
Copyright © 2021 Xie, Luo, He and Tang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
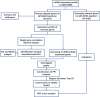
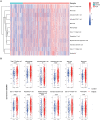


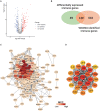
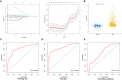
References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

