Biophysical Compatibility of a Heterotrimeric Tyrosinase-TYRP1-TYRP2 Metalloenzyme Complex
- PMID: 33995009
- PMCID: PMC8114058
- DOI: 10.3389/fphar.2021.602206
Biophysical Compatibility of a Heterotrimeric Tyrosinase-TYRP1-TYRP2 Metalloenzyme Complex
Abstract
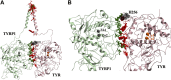
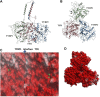
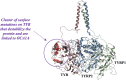
Tyrosinase (TYR) is a copper-containing monooxygenase central to the function of melanocytes. Alterations in its expression or activity contribute to variations in skin, hair and eye color, and underlie a variety of pathogenic pigmentary phenotypes, including several forms of oculocutaneous albinism (OCA). Many of these phenotypes are linked to individual missense mutations causing single nucleotide variants and polymorphisms (SNVs) in TYR. We previously showed that two TYR homologues, TYRP1 and TYRP2, modulate TYR activity and stabilize the TYR protein. Accordingly, to investigate whether TYR, TYRP1, and TYRP2 are biophysically compatible with various heterocomplexes, we computationally docked a high-quality 3D model of TYR to the crystal structure of TYRP1 and to a high-quality 3D model of TYRP2. Remarkably, the resulting TYR-TYRP1 heterodimer was complementary in structure and energy with the TYR-TYRP2 heterodimer, with TYRP1 and TYRP2 docking to different adjacent surfaces on TYR that apposed a third realistic protein interface between TYRP1-TYRP2. Hence, the 3D models are compatible with a heterotrimeric TYR-TYRP1-TYRP2 complex. In addition, this heterotrimeric TYR-TYRP1-TYRP2 positioned the C-terminus of each folded enzymatic domain in an ideal position to allow their C-terminal transmembrane helices to form a putative membrane embedded three-helix bundle. Finally, pathogenic TYR mutations causing OCA1A, which also destabilize TYR biochemically, cluster on an unoccupied protein interface at the periphery of the heterotrimeric complex, suggesting that this may be a docking site for OCA2, an anion channel. Pathogenic OCA2 mutations result in similar phenotypes to those produced by OCA1A TYR mutations. While this complex may be difficult to detect in vitro, due to the complex environment of the vertebrate cellular membranous system, our results support the existence of a heterotrimeric complex in melanogenesis.
Keywords: computational molecular docking; melanosome; molecular modeling; oculocutaneous albinism; pigmentary disorders; protein-protein interface; tyrosinase.
Copyright © 2021 Lavinda, Manga, Orlow and Cardozo.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



Similar articles
-
SLC45A2 mutation frequency in Oculocutaneous Albinism Italian patients doesn't differ from other European studies.Gene. 2014 Jan 1;533(1):398-402. doi: 10.1016/j.gene.2013.09.053. Epub 2013 Oct 3. Gene. 2014. PMID: 24096233
-
Comparable down-regulation of TYR, TYRP1 and TYRP2 genes and inhibition of melanogenesis by tyrostat, tocotrienol-rich fraction and tocopherol in human skin melanocytes improves skin pigmentation.Clin Ter. 2014;165(1):e39-45. Clin Ter. 2014. PMID: 24589959
-
Membrane transport proteins in melanosomes: Regulation of ions for pigmentation.Biochim Biophys Acta Biomembr. 2020 Dec 1;1862(12):183318. doi: 10.1016/j.bbamem.2020.183318. Epub 2020 Apr 22. Biochim Biophys Acta Biomembr. 2020. PMID: 32333855 Free PMC article. Review.
-
Tyrosinase and ocular diseases: some novel thoughts on the molecular basis of oculocutaneous albinism type 1.Prog Retin Eye Res. 2007 Jul;26(4):323-58. doi: 10.1016/j.preteyeres.2007.01.001. Epub 2007 Jan 17. Prog Retin Eye Res. 2007. PMID: 17355913 Review.
-
Chromosomal structure of the human TYRP1 and TYRP2 loci and comparison of the tyrosinase-related protein gene family.Genomics. 1995 Sep 1;29(1):24-34. doi: 10.1006/geno.1995.1211. Genomics. 1995. PMID: 8530077
Cited by
-
Establishment of a synchronized tyrosinase transport system revealed a role of Tyrp1 in efficient melanogenesis by promoting tyrosinase targeting to melanosomes.Sci Rep. 2024 Jan 30;14(1):2529. doi: 10.1038/s41598-024-53072-6. Sci Rep. 2024. PMID: 38291221 Free PMC article.
-
Theoretical Studies of Cyanophycin Dipeptides as Inhibitors of Tyrosinases.Int J Mol Sci. 2022 Mar 19;23(6):3335. doi: 10.3390/ijms23063335. Int J Mol Sci. 2022. PMID: 35328756 Free PMC article.
-
Protein Biochemistry and Molecular Modeling of the Intra-Melanosomal Domain of Human Recombinant Tyrp2 Protein and OCA8-Related Mutant Variants.Int J Mol Sci. 2022 Jan 24;23(3):1305. doi: 10.3390/ijms23031305. Int J Mol Sci. 2022. PMID: 35163231 Free PMC article.
-
Genetic variants in melanogenesis proteins TYRP1 and TYR are associated with the golden rhesus macaque phenotype.G3 (Bethesda). 2023 Sep 30;13(10):jkad168. doi: 10.1093/g3journal/jkad168. G3 (Bethesda). 2023. PMID: 37522525 Free PMC article.
-
Drug Repurposing of Voglibose, a Diabetes Medication for Skin Health.Pharmaceuticals (Basel). 2025 Feb 7;18(2):224. doi: 10.3390/ph18020224. Pharmaceuticals (Basel). 2025. PMID: 40006038 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

