Proteomics and Machine Learning Approaches Reveal a Set of Prognostic Markers for COVID-19 Severity With Drug Repurposing Potential
- PMID: 33995121
- PMCID: PMC8120435
- DOI: 10.3389/fphys.2021.652799
Proteomics and Machine Learning Approaches Reveal a Set of Prognostic Markers for COVID-19 Severity With Drug Repurposing Potential
Abstract
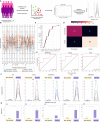
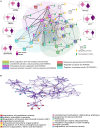
The pestilential pathogen SARS-CoV-2 has led to a seemingly ceaseless pandemic of COVID-19. The healthcare sector is under a tremendous burden, thus necessitating the prognosis of COVID-19 severity. This in-depth study of plasma proteome alteration provides insights into the host physiological response towards the infection and also reveals the potential prognostic markers of the disease. Using label-free quantitative proteomics, we performed deep plasma proteome analysis in a cohort of 71 patients (20 COVID-19 negative, 18 COVID-19 non-severe, and 33 severe) to understand the disease dynamics. Of the 1200 proteins detected in the patient plasma, 38 proteins were identified to be differentially expressed between non-severe and severe groups. The altered plasma proteome revealed significant dysregulation in the pathways related to peptidase activity, regulated exocytosis, blood coagulation, complement activation, leukocyte activation involved in immune response, and response to glucocorticoid biological processes in severe cases of SARS-CoV-2 infection. Furthermore, we employed supervised machine learning (ML) approaches using a linear support vector machine model to identify the classifiers of patients with non-severe and severe COVID-19. The model used a selected panel of 20 proteins and classified the samples based on the severity with a classification accuracy of 0.84. Putative biomarkers such as angiotensinogen and SERPING1 and ML-derived classifiers including the apolipoprotein B, SERPINA3, and fibrinogen gamma chain were validated by targeted mass spectrometry-based multiple reaction monitoring (MRM) assays. We also employed an in silico screening approach against the identified target proteins for the therapeutic management of COVID-19. We shortlisted two FDA-approved drugs, namely, selinexor and ponatinib, which showed the potential of being repurposed for COVID-19 therapeutics. Overall, this is the first most comprehensive plasma proteome investigation of COVID-19 patients from the Indian population, and provides a set of potential biomarkers for the disease severity progression and targets for therapeutic interventions.
Keywords: COVID-19 plasma; drug-repurposing; host response; machine learning; mass spectrometry; molecular pathways; prognostic biomarkers; proteomics.
Copyright © 2021 Suvarna, Biswas, Pai, Acharjee, Bankar, Palanivel, Salkar, Verma, Mukherjee, Choudhury, Ghantasala, Ghosh, Singh, Banerjee, Badaya, Bihani, Loya, Mantri, Burli, Roy, Srivastava, Agrawal, Shrivastav, Shastri and Srivastava.
Conflict of interest statement
The authors have filed an Indian patent related to this work “Protein markers and method for prognosis of COVID-19 in individuals” (Application number: 202023054753). The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
-
- Alsuliman T., Humaidan D., Sliman L. (2020). Machine learning and artificial intelligence in the service of medicine: necessity or potentiality?. Curr. Res. Transl. Med. 68 245–251. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

