Biphenyl 2,3-Dioxygenase in Pseudomonas alcaliphila JAB1 Is Both Induced by Phenolics and Monoterpenes and Involved in Their Transformation
- PMID: 33995321
- PMCID: PMC8119895
- DOI: 10.3389/fmicb.2021.657311
Biphenyl 2,3-Dioxygenase in Pseudomonas alcaliphila JAB1 Is Both Induced by Phenolics and Monoterpenes and Involved in Their Transformation
Abstract
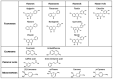
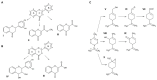
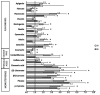
The involvement of bacterial aromatic ring-hydroxylating dioxygenases (ARHDs) in the degradation of aromatic pollutants, such as polychlorinated biphenyls (PCBs), has been well studied. However, there is considerable speculation as to the origin of this ability. One hypothesis is centered on a connection between the ability to degrade aromatic pollutants and the necessity of soil bacteria to cope with and/or utilize secondary plant metabolites (SPMs). To investigate this connection, we researched the involvement of biphenyl 2,3-dioxygenase (BPDO), an ARHD essential for the degradation of PCBs, in the metabolism of SPMs in the soil bacterium Pseudomonas alcaliphila JAB1, a versatile degrader of PCBs. We demonstrated the ability of the strain JAB1 to transform a variety of SPMs, namely the flavonoids apigenin, flavone, flavanone, naringenin, fisetin, quercetin, morin, and catechin, caffeic acid, trans-cinnamic acid, and the monoterpenes (S)-limonene and (R)-carvone. Of those, the transformation of flavone, flavanone, and (S)-limonene was conditioned by the activity of JAB1-borne BPDO and thus was researched in more detail, and we found evidence for the limonene monooxygenase activity of the BPDO. Furthermore, the bphA gene in the strain JAB1 was demonstrated to be induced by a wide range of SPMs, with monoterpenes being the strongest inducers of the SPMs tested. Thus, our findings contribute to the growing body of evidence that ARHDs not only play a role in the catabolism of aromatic pollutants, but also of natural plant-derived aromatics, and this study supports the hypothesis that ARHDs participate in ecological processes mediated by SPMs.
Keywords: aromatic ring-hydroxylating dioxygenases; biphenyl dioxygenase; monoterpenes; phenolics; secondary plant metabolites.
Copyright © 2021 Zubrova, Michalikova, Semerad, Strejcek, Cajthaml, Suman and Uhlik.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Differential effect of monoterpenes and flavonoids on the transcription of aromatic ring-hydroxylating dioxygenase genes in Rhodococcus opacus C1 and Rhodococcus sp. WAY2.Microb Genom. 2025 Mar;11(3):001359. doi: 10.1099/mgen.0.001359. Microb Genom. 2025. PMID: 40042991 Free PMC article.
-
Predominant Biphenyl Dioxygenase From Legacy Polychlorinated Biphenyl (PCB)-Contaminated Soil Is a Part of Unusual Gene Cluster and Transforms Flavone and Flavanone.Front Microbiol. 2021 Oct 14;12:644708. doi: 10.3389/fmicb.2021.644708. eCollection 2021. Front Microbiol. 2021. PMID: 34721309 Free PMC article.
-
Complete genome sequence of Pseudomonas alcaliphila JAB1 (=DSM 26533), a versatile degrader of organic pollutants.Stand Genomic Sci. 2018 Feb 1;13:3. doi: 10.1186/s40793-017-0306-7. eCollection 2018. Stand Genomic Sci. 2018. PMID: 29435100 Free PMC article.
-
Biphenyl-utilizing bacteria and their functional genes in a pine root zone contaminated with polychlorinated biphenyls (PCBs).ISME J. 2007 Jun;1(2):134-48. doi: 10.1038/ismej.2007.26. Epub 2007 May 24. ISME J. 2007. PMID: 18043623
-
Flavonoids biotransformation by bacterial non-heme dioxygenases, biphenyl and naphthalene dioxygenase.Appl Microbiol Biotechnol. 2011 Jul;91(2):219-28. doi: 10.1007/s00253-011-3334-z. Epub 2011 May 28. Appl Microbiol Biotechnol. 2011. PMID: 21626021 Review.
Cited by
-
Polychlorinated biphenyls modify Arabidopsis root exudation pattern to accommodate degrading bacteria, showing strain and functional trait specificity.Front Plant Sci. 2024 Jul 5;15:1429096. doi: 10.3389/fpls.2024.1429096. eCollection 2024. Front Plant Sci. 2024. PMID: 39036359 Free PMC article.
-
Differential effect of monoterpenes and flavonoids on the transcription of aromatic ring-hydroxylating dioxygenase genes in Rhodococcus opacus C1 and Rhodococcus sp. WAY2.Microb Genom. 2025 Mar;11(3):001359. doi: 10.1099/mgen.0.001359. Microb Genom. 2025. PMID: 40042991 Free PMC article.
-
Evolution of genetic architecture and gene regulation in biphenyl/PCB-degrading bacteria.Front Microbiol. 2023 Jun 7;14:1168246. doi: 10.3389/fmicb.2023.1168246. eCollection 2023. Front Microbiol. 2023. PMID: 37350784 Free PMC article. Review.
-
Predominant Biphenyl Dioxygenase From Legacy Polychlorinated Biphenyl (PCB)-Contaminated Soil Is a Part of Unusual Gene Cluster and Transforms Flavone and Flavanone.Front Microbiol. 2021 Oct 14;12:644708. doi: 10.3389/fmicb.2021.644708. eCollection 2021. Front Microbiol. 2021. PMID: 34721309 Free PMC article.
-
Flavonoids Are Intra- and Inter-Kingdom Modulator Signals.Microorganisms. 2022 Dec 15;10(12):2479. doi: 10.3390/microorganisms10122479. Microorganisms. 2022. PMID: 36557733 Free PMC article. Review.
References
-
- Agullo L., Romero-Silva M. J., Domenech M., Seeger M. (2017). p-cymene promotes its catabolism through the p-cymene and the p-cumate pathways, activates a stress response and reduces the biofilm formation in Burkholderia xenovorans LB400. PLoS One 12:e0169544. 10.1371/journal.pone.0169544, PMID: - DOI - PMC - PubMed
-
- Aoki H., Kimura T., Habe H., Yamane H., Kodama T., Omori T. (1996). Cloning, nucleotide sequence, and characterization of the genes encoding enzymes involved in the degradation of cumene to 2-hydroxy-6-oxo-7-methylocta-2,4-dienoic acid in Pseudomonas fluorescens IP01. J. Ferment. Bioeng. 81, 187–196. 10.1016/0922-338X(96)82207-0 - DOI
-
- Benjamini Y., Hochberg Y. (1995). Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Ser. B Methodol. 57, 289–300. 10.1111/j.2517-6161.1995.tb02031.x - DOI
-
- Böttger A., Vothknecht U., Bolle C., Wolf A. (eds.) (2018). “Plant Secondary Metabolites and Their General Function in Plants,” in Lessons on Caffeine, Cannabis & Co: Plant-derived Drugs and Their Interaction With Human Receptors. (Cham: Springer International Publishing; ), 3–17.
LinkOut - more resources
Full Text Sources
Other Literature Sources

