How Microbes Defend Themselves From Incoming Hydrogen Peroxide
- PMID: 33995399
- PMCID: PMC8115020
- DOI: 10.3389/fimmu.2021.667343
How Microbes Defend Themselves From Incoming Hydrogen Peroxide
Abstract
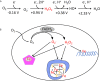
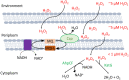
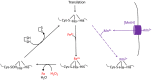
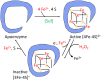
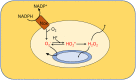
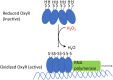
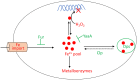
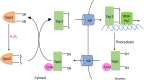
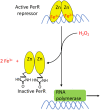
Microbes rely upon iron as a cofactor for many enzymes in their central metabolic processes. The reactive oxygen species (ROS) superoxide and hydrogen peroxide react rapidly with iron, and inside cells they can generate both enzyme and DNA damage. ROS are formed in some bacterial habitats by abiotic processes. The vulnerability of bacteria to ROS is also apparently exploited by ROS-generating host defense systems and bacterial competitors. Phagocyte-derived can toxify captured bacteria by damaging unidentified biomolecules on the cell surface; it is unclear whether phagocytic H2O2, which can penetrate into the cell interior, also plays a role in suppressing bacterial invasion. Both pathogenic and free-living microbes activate defensive strategies to defend themselves against incoming H2O2. Most bacteria sense the H2O2via OxyR or PerR transcription factors, whereas yeast uses the Grx3/Yap1 system. In general these regulators induce enzymes that reduce cytoplasmic H2O2 concentrations, decrease the intracellular iron pools, and repair the H2O2-mediated damage. However, individual organisms have tailored these transcription factors and their regulons to suit their particular environmental niches. Some bacteria even contain both OxyR and PerR, raising the question as to why they need both systems. In lab experiments these regulators can also respond to nitric oxide and disulfide stress, although it is unclear whether the responses are physiologically relevant. The next step is to extend these studies to natural environments, so that we can better understand the circumstances in which these systems act. In particular, it is important to probe the role they may play in enabling host infection by microbial pathogens.
Keywords: OxyR regulator; Yap1p; nitric oxide; peroxide sensing repressor (PerR); reactive oxygen species.
Copyright © 2021 Sen and Imlay.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

