Structural and molecular biology of hepatitis E virus
- PMID: 33995894
- PMCID: PMC8079827
- DOI: 10.1016/j.csbj.2021.03.038
Structural and molecular biology of hepatitis E virus
Abstract
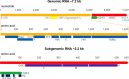
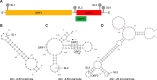
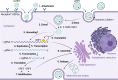
Hepatitis E virus (HEV) is one of the most common causes of acute viral hepatitis, mainly transmitted by fecal-oral route but has also been linked to fulminant hepatic failure, chronic hepatitis, and extrahepatic neurological and renal diseases. HEV is an emerging zoonotic pathogen with a broad host range, and strains of HEV from numerous animal species are known to cross species barriers and infect humans. HEV is a single-stranded, positive-sense RNA virus in the family Hepeviridae. The genome typically contains three open reading frames (ORFs): ORF1 encodes a nonstructural polyprotein for virus replication and transcription, ORF2 encodes the capsid protein that elicits neutralizing antibodies, and ORF3, which partially overlaps ORF2, encodes a multifunctional protein involved in virion morphogenesis and pathogenesis. HEV virions are non-enveloped spherical particles in feces but exist as quasi-enveloped particles in circulating blood. Two types of HEV virus-like particles (VLPs), small T = 1 (270 Å) and native virion-sized T = 3 (320-340 Å) have been reported. There exist two distinct forms of capsid protein, the secreted form (ORF2S) inhibits antibody neutralization, whereas the capsid-associated form (ORF2C) self-assembles to VLPs. Four cis-reactive elements (CREs) containing stem-loops from secondary RNA structures have been identified in the non-coding regions and are critical for virus replication. This mini-review discusses the current knowledge and gaps regarding the structural and molecular biology of HEV with emphasis on the virion structure, genomic organization, secondary RNA structures, viral proteins and their functions, and life cycle of HEV.
Keywords: Genetic Diversity; Genomic organization; Hepatitis E Virus (HEV); Life cycle of HEV; Proteins and functions; Virion structure.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures


References
-
- Rein D.B., Stevens G.A., Theaker J., Wittenborn J.S., Wiersma S.T. The global burden of hepatitis E virus genotypes 1 and 2 in 2005. Hepatology. 2012;55:988–997. - PubMed
-
- Perez-Gracia M.T., Suay-Garcia B., Mateos-Lindemann M.L. Hepatitis E and pregnancy: current state. Rev Med Virol. 2017;27 - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
