DRscDB: A single-cell RNA-seq resource for data mining and data comparison across species
- PMID: 33995899
- PMCID: PMC8085783
- DOI: 10.1016/j.csbj.2021.04.021
DRscDB: A single-cell RNA-seq resource for data mining and data comparison across species
Abstract
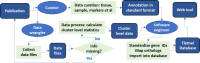
With the advent of single-cell RNA sequencing (scRNA-seq) technologies, there has been a spike in studies involving scRNA-seq of several tissues across diverse species including Drosophila. Although a few databases exist for users to query genes of interest within the scRNA-seq studies, search tools that enable users to find orthologous genes and their cell type-specific expression patterns across species are limited. Here, we built a new search database, DRscDB (https://www.flyrnai.org/tools/single_cell/web/), to address this need. DRscDB serves as a comprehensive repository for published scRNA-seq datasets for Drosophila and relevant datasets from human and other model organisms. DRscDB is based on manual curation of Drosophila scRNA-seq studies of various tissue types and their corresponding analogous tissues in vertebrates including zebrafish, mouse, and human. Of note, our search database provides most of the literature-derived marker genes, thus preserving the original analysis of the published scRNA-seq datasets. Finally, DRscDB serves as a web-based user interface that allows users to mine gene expression data from scRNA-seq studies and perform cell cluster enrichment analyses pertaining to various scRNA-seq studies, both within and across species.
Keywords: Cross-species analysis; Data mining; Model organisms; single-cell RNA-seq.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




Similar articles
-
CellDepot: A Unified Repository for scRNA-seq Data and Visual Exploration.J Mol Biol. 2022 Jun 15;434(11):167425. doi: 10.1016/j.jmb.2021.167425. Epub 2021 Dec 28. J Mol Biol. 2022. PMID: 34971674
-
scRNASeqDB: A Database for RNA-Seq Based Gene Expression Profiles in Human Single Cells.Genes (Basel). 2017 Dec 5;8(12):368. doi: 10.3390/genes8120368. Genes (Basel). 2017. PMID: 29206167 Free PMC article.
-
Exploring the single-cell RNA-seq analysis landscape with the scRNA-tools database.PLoS Comput Biol. 2018 Jun 25;14(6):e1006245. doi: 10.1371/journal.pcbi.1006245. eCollection 2018 Jun. PLoS Comput Biol. 2018. PMID: 29939984 Free PMC article.
-
A Roadmap for Selecting and Utilizing Optimal Features in scRNA Sequencing Data Analysis for Stem Cell Research: A Comprehensive Review.Int J Stem Cells. 2024 Nov 30;17(4):347-362. doi: 10.15283/ijsc23170. Epub 2024 Mar 27. Int J Stem Cells. 2024. PMID: 38531607 Free PMC article. Review.
-
Single-cell RNA sequencing in breast cancer: Understanding tumor heterogeneity and paving roads to individualized therapy.Cancer Commun (Lond). 2020 Aug;40(8):329-344. doi: 10.1002/cac2.12078. Epub 2020 Jul 12. Cancer Commun (Lond). 2020. PMID: 32654419 Free PMC article. Review.
Cited by
-
Glucocerebrosidase deficiency leads to neuropathology via cellular immune activation.bioRxiv [Preprint]. 2023 Dec 13:2023.12.13.571406. doi: 10.1101/2023.12.13.571406. bioRxiv. 2023. Update in: PLoS Genet. 2024 Nov 11;20(11):e1011105. doi: 10.1371/journal.pgen.1011105. PMID: 38168223 Free PMC article. Updated. Preprint.
-
Molecular traces of Drosophila hemocytes reveal transcriptomic conservation with vertebrate myeloid cells.PLoS Genet. 2023 Dec 19;19(12):e1011077. doi: 10.1371/journal.pgen.1011077. eCollection 2023 Dec. PLoS Genet. 2023. PMID: 38113249 Free PMC article.
-
Single-cell analysis of mosquito hemocytes identifies signatures of immune cell subtypes and cell differentiation.Elife. 2021 Jul 28;10:e66192. doi: 10.7554/eLife.66192. Elife. 2021. PMID: 34318744 Free PMC article.
-
A pleiotropic chemoreceptor facilitates the production and perception of mating pheromones.iScience. 2022 Dec 29;26(1):105882. doi: 10.1016/j.isci.2022.105882. eCollection 2023 Jan 20. iScience. 2022. PMID: 36691619 Free PMC article.
-
Transcriptional programs mediating neuronal toxicity and altered glial-neuronal signaling in a Drosophila knock-in tauopathy model.Genome Res. 2024 May 15;34(4):590-605. doi: 10.1101/gr.278576.123. Genome Res. 2024. PMID: 38599684 Free PMC article.
References
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
