Experimental approaches for investigating ion atmospheres around nucleic acids and proteins
- PMID: 33995919
- PMCID: PMC8102144
- DOI: 10.1016/j.csbj.2021.04.033
Experimental approaches for investigating ion atmospheres around nucleic acids and proteins
Abstract
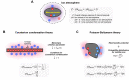
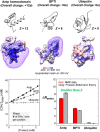
Ionic interactions are crucial to biological functions of DNA, RNA, and proteins. Experimental research on how ions behave around biological macromolecules has lagged behind corresponding theoretical and computational research. In the 21st century, quantitative experimental approaches for investigating ionic interactions of biomolecules have become available and greatly facilitated examinations of theoretical electrostatic models. These approaches utilize anomalous small-angle X-ray scattering, atomic emission spectroscopy, mass spectrometry, or nuclear magnetic resonance (NMR) spectroscopy. We provide an overview on the experimental methodologies that can quantify and characterize ions within the ion atmospheres around nucleic acids, proteins, and their complexes.
Keywords: AES, atomic emission spectroscopy; ASAXS, anomalous small-angle X-ray scattering; BE, buffer equilibration; Dynamics; Electrostatic interactions; ICP-MS, inductively coupled plasma mass spectrometry; Ion atmosphere; Ionic diffusion; NMR, nuclear magnetic resonance; PRE, paramagnetic relaxation enhancement; SAXS, small-angle X-ray scattering; Spatial distribution.
© 2021 The Author(s).
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures



Similar articles
-
Counting ions around DNA with anomalous small-angle X-ray scattering.J Am Chem Soc. 2010 Nov 24;132(46):16334-6. doi: 10.1021/ja107259y. Epub 2010 Nov 3. J Am Chem Soc. 2010. PMID: 21047071 Free PMC article.
-
Using anomalous small angle X-ray scattering to probe the ion atmosphere around nucleic acids.Methods Enzymol. 2009;469:391-410. doi: 10.1016/S0076-6879(09)69019-4. Epub 2009 Nov 17. Methods Enzymol. 2009. PMID: 20946800
-
Dynamics of Ionic Interactions at Protein-Nucleic Acid Interfaces.Acc Chem Res. 2020 Sep 15;53(9):1802-1810. doi: 10.1021/acs.accounts.0c00212. Epub 2020 Aug 26. Acc Chem Res. 2020. PMID: 32845610 Free PMC article.
-
SAXS studies of ion-nucleic acid interactions.Annu Rev Biophys. 2011;40:225-42. doi: 10.1146/annurev-biophys-042910-155349. Annu Rev Biophys. 2011. PMID: 21332357 Review.
-
Molecular Dynamics Simulations Combined with Nuclear Magnetic Resonance and/or Small-Angle X-ray Scattering Data for Characterizing Intrinsically Disordered Protein Conformational Ensembles.J Chem Inf Model. 2019 May 28;59(5):1743-1758. doi: 10.1021/acs.jcim.8b00928. Epub 2019 Mar 18. J Chem Inf Model. 2019. PMID: 30840442 Review.
Cited by
-
RNA Electrostatics: How Ribozymes Engineer Active Sites to Enable Catalysis.J Phys Chem B. 2022 Aug 18;126(32):5982-5990. doi: 10.1021/acs.jpcb.2c03727. Epub 2022 Jul 21. J Phys Chem B. 2022. PMID: 35862934 Free PMC article.
-
Ion NMR for Biomolecular Systems.J Mol Biol. 2025 Jun 6:169285. doi: 10.1016/j.jmb.2025.169285. Online ahead of print. J Mol Biol. 2025. PMID: 40484343 Review.
-
Sensing the structural and conformational properties of single-stranded nucleic acids using electrometry and molecular simulations.Sci Rep. 2024 Sep 4;14(1):20582. doi: 10.1038/s41598-024-70641-x. Sci Rep. 2024. PMID: 39232063 Free PMC article.
-
Dynamics of Cations around DNA and Protein as Revealed by 23Na Diffusion NMR Spectroscopy.Anal Chem. 2022 Feb 8;94(5):2444-2452. doi: 10.1021/acs.analchem.1c04197. Epub 2022 Jan 26. Anal Chem. 2022. PMID: 35080384 Free PMC article.
-
Measuring Local Electrostatic Potentials Around Nucleic Acids by Paramagnetic NMR Spectroscopy.J Phys Chem Lett. 2022 Oct 27;13(42):10025-10029. doi: 10.1021/acs.jpclett.2c02623. Epub 2022 Oct 20. J Phys Chem Lett. 2022. PMID: 36264151 Free PMC article.
References
-
- Okur H.I., Hladílková J., Rembert K.B., Cho Y., Heyda J., Dzubiella J. Beyond the Hofmeister Series: Ion-Specific Effects on Proteins and Their Biological Functions. J Phys Chem B. 2017;121(9):1997–2014. - PubMed
-
- Record M.T., Jr., Zhang W., Anderson C.F. Analysis of effects of salts and uncharged solutes on protein and nucleic acid equilibria and processes: a practical guide to recognizing and interpreting polyelectrolyte effects, Hofmeister effects, and osmotic effects of salts. Adv Protein Chem. 1998;51:281–353. - PubMed
-
- Zhang Y., Cremer P.S. Chemistry of Hofmeister anions and osmolytes. Annu Rev Phys Chem. 2010;61(1):63–83. - PubMed
Publication types
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
