Identification of Differentially Expressed Human Endogenous Retrovirus Families in Human Leukemia and Lymphoma Cell Lines and Stem Cells
- PMID: 33996550
- PMCID: PMC8117144
- DOI: 10.3389/fonc.2021.637981
Identification of Differentially Expressed Human Endogenous Retrovirus Families in Human Leukemia and Lymphoma Cell Lines and Stem Cells
Abstract
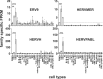
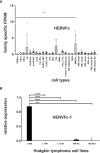
Endogenous retroviruses (ERVs) are becoming more and more relevant in cancer research and might be potential targets. The oncogenic potential of human ERVs (HERVs) has been recognized and includes immunosuppression, cell fusion, antigenicity of viral proteins, and regulation of neighboring genes. To decipher the role of HERVs in human cancers, we used a bioinformatics approach and analyzed RNA sequencing data from the LL-100 panel, covering 22 entities of hematopoietic neoplasias including T cell, B cell and myeloid malignancies. We compared HERV expression in this panel with hematopoietic stem cells (HSCs), embryonic stem cells (ESCs) and normal blood cells. RNA sequencing data were mapped against a comprehensive synthetic viral metagenome with 116 HERV sequences from 14 different HERV families. Of these, 13 HERV families and elements were differently expressed in malignant hematopoietic cells and stem cells. We found transcriptional upregulation of HERVE family in acute megakaryocytic and erythroid leukemia and of HERVFc family in multiple myeloma/plasma cell leukemia (PCL). The HERVFc member HERVFc-1 was found transcriptionally active in the multiple myeloma cell line OPM-2 and also in the Hodgkin lymphoma cell line L-428. The expression of HERVFc-1 in L-428 cells was validated by qRT-PCR. We also confirm transcriptional downregulation of ERV3 in acute megakaryocytic and erythroid leukemia, and HERVK in acute monocytic and myelocytic leukemia and a depression of HERVF in all malignant entities. Most of the higher expressed HERV families could be detected in stem cells including HERVK (HML-2), HERV-like, HERVV, HERVT, ERV9, HERVW, HERVF, HERVMER, ERV3, HERVH and HERVPABLB.
Keywords: LL-100 panel; RNA sequencing; embryonic stem cells (ESCs); gene expression; hematopoietic stem cells (HSCs); human endogenous retroviruses (HERVs); leukemia; lymphoma.
Copyright © 2021 Engel, Wieland, Krüger, Volkmer, Cynis, Emmer and Staege.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures



References
LinkOut - more resources
Full Text Sources
Other Literature Sources

