The Unfolded Protein Response and Autophagy on the Crossroads of Coronaviruses Infections
- PMID: 33996638
- PMCID: PMC8113818
- DOI: 10.3389/fcimb.2021.668034
The Unfolded Protein Response and Autophagy on the Crossroads of Coronaviruses Infections
Abstract
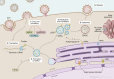
The ability to sense and adequately respond to variable environmental conditions is central for cellular and organismal homeostasis. Eukaryotic cells are equipped with highly conserved stress-response mechanisms that support cellular function when homeostasis is compromised, promoting survival. Two such mechanisms - the unfolded protein response (UPR) and autophagy - are involved in the cellular response to perturbations in the endoplasmic reticulum, in calcium homeostasis, in cellular energy or redox status. Each of them operates through conserved signaling pathways to promote cellular adaptations that include re-programming transcription of genes and translation of new proteins and degradation of cellular components. In addition to their specific functions, it is becoming increasingly clear that these pathways intersect in many ways in different contexts of cellular stress. Viral infections are a major cause of cellular stress as many cellular functions are coopted to support viral replication. Both UPR and autophagy are induced upon infection with many different viruses with varying outcomes - in some instances controlling infection while in others supporting viral replication and infection. The role of UPR and autophagy in response to coronavirus infection has been a matter of debate in the last decade. It has been suggested that CoV exploit components of autophagy machinery and UPR to generate double-membrane vesicles where it establishes its replicative niche and to control the balance between cell death and survival during infection. Even though the molecular mechanisms are not fully elucidated, it is clear that UPR and autophagy are intimately associated during CoV infections. The current SARS-CoV-2 pandemic has brought renewed interest to this topic as several drugs known to modulate autophagy - including chloroquine, niclosamide, valinomycin, and spermine - were proposed as therapeutic options. Their efficacy is still debatable, highlighting the need to better understand the molecular interactions between CoV, UPR and autophagy.
Keywords: autophagy; coronavirus; host-pathogen interaction; integrated stress response; unfolded protein response.
Copyright © 2021 Prestes, Bruno, Travassos and Carneiro.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
SARS-CoV-2 Diverges from Other Betacoronaviruses in Only Partially Activating the IRE1α/XBP1 Endoplasmic Reticulum Stress Pathway in Human Lung-Derived Cells.mBio. 2022 Oct 26;13(5):e0241522. doi: 10.1128/mbio.02415-22. Epub 2022 Sep 20. mBio. 2022. PMID: 36125275 Free PMC article.
-
P.1 and P.2 SARS-CoV-2 Brazilian variants activate the unfolded protein response with a time and pathway specificity.J Proteomics. 2025 May 15;315:105397. doi: 10.1016/j.jprot.2025.105397. Epub 2025 Feb 3. J Proteomics. 2025. PMID: 39909104
-
Autophagy, Unfolded Protein Response, and Neuropilin-1 Cross-Talk in SARS-CoV-2 Infection: What Can Be Learned from Other Coronaviruses.Int J Mol Sci. 2021 Jun 1;22(11):5992. doi: 10.3390/ijms22115992. Int J Mol Sci. 2021. PMID: 34206057 Free PMC article. Review.
-
The PERK/PKR-eIF2α Pathway Negatively Regulates Porcine Hemagglutinating Encephalomyelitis Virus Replication by Attenuating Global Protein Translation and Facilitating Stress Granule Formation.J Virol. 2022 Jan 12;96(1):e0169521. doi: 10.1128/JVI.01695-21. Epub 2021 Oct 13. J Virol. 2022. PMID: 34643429 Free PMC article.
-
The roles and mechanisms of endoplasmic reticulum stress-mediated autophagy in animal viral infections.Vet Res. 2024 Sep 3;55(1):107. doi: 10.1186/s13567-024-01360-4. Vet Res. 2024. PMID: 39227990 Free PMC article. Review.
Cited by
-
Wheat Germ Spermidine and Clove Eugenol in Combination Stimulate Autophagy In Vitro Showing Potential in Supporting the Immune System against Viral Infections.Molecules. 2022 May 26;27(11):3425. doi: 10.3390/molecules27113425. Molecules. 2022. PMID: 35684363 Free PMC article.
-
Insights into the Activation of Unfolded Protein Response Mechanism during Coronavirus Infection.Curr Issues Mol Biol. 2024 May 5;46(5):4286-4308. doi: 10.3390/cimb46050261. Curr Issues Mol Biol. 2024. PMID: 38785529 Free PMC article. Review.
-
Spring viraemia of carp virus modulates the time-dependent unfolded protein response to facilitate viral replication.Front Immunol. 2025 Apr 3;16:1576758. doi: 10.3389/fimmu.2025.1576758. eCollection 2025. Front Immunol. 2025. PMID: 40248709 Free PMC article.
-
SARS-CoV-2 Diverges from Other Betacoronaviruses in Only Partially Activating the IRE1α/XBP1 Endoplasmic Reticulum Stress Pathway in Human Lung-Derived Cells.mBio. 2022 Oct 26;13(5):e0241522. doi: 10.1128/mbio.02415-22. Epub 2022 Sep 20. mBio. 2022. PMID: 36125275 Free PMC article.
-
Mantis: High-throughput 4D imaging and analysis of the molecular and physical architecture of cells.PNAS Nexus. 2024 Aug 9;3(9):pgae323. doi: 10.1093/pnasnexus/pgae323. eCollection 2024 Sep. PNAS Nexus. 2024. PMID: 39282007 Free PMC article.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Miscellaneous

