Peptidyl Activity-Based Probes for Imaging Serine Proteases
- PMID: 33996745
- PMCID: PMC8117214
- DOI: 10.3389/fchem.2021.639410
Peptidyl Activity-Based Probes for Imaging Serine Proteases
Abstract
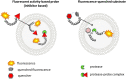
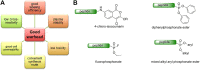
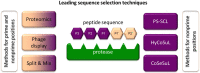
Proteases catalyze the hydrolysis of peptide bonds. Products of this breakdown mediate signaling in an enormous number of biological processes. Serine proteases constitute the most numerous group of proteases, accounting for 40%, and they are prevalent in many physiological functions, both normal and disease-related functions, making them one of the most important enzymes in humans. The activity of proteases is controlled at the expression level by posttranslational modifications and/or endogenous inhibitors. The study of serine proteases requires specific reagents not only for detecting their activity but also for their imaging. Such tools include inhibitors or substrate-related chemical molecules that allow the detection of proteolysis and visual observation of active enzymes, thus facilitating the characterization of the activity of proteases in the complex proteome. Peptidyl activity-based probes (ABPs) have been extensively studied recently, and this review describes the basic principles in the design of peptide-based imaging agents for serine proteases, provides examples of activity-based probe applications and critically discusses their strengths, weaknesses, challenges and limitations.
Keywords: activity-based probes; chemical reagents; enzyme detection; imaging; internally quenched fluorogenic substrates; serine proteases.
Copyright © 2021 Kasperkiewicz.
Conflict of interest statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




References
-
- Aaltonen N., Singha P. K., Jakupovic H., Wirth T., Samaranayake H., Pasonen-Seppanen S., et al. (2020). High-Resolution Confocal Fluorescence Imaging of Serine Hydrolase Activity in Cryosections—Application to Glioma Brain Unveils Activity Hotspots Originating from Tumor-Associated Neutrophils. Biol. Proced. Online. 22, 6. 10.1186/s12575-020-00118-4 - DOI - PMC - PubMed
-
- Bender K. O., Ofori L., van der Linden W. A., Mock E. D., Datta G. K., Chowdhury S., et al. (2015). Design of a Highly Selective Quenched Activity-Based Probe and its Application in Dual Color Imaging Studies of Cathepsin S Activity Localization. J. Am. Chem. Soc. 137 (14), 4771–4777. 10.1021/jacs.5b00315 - DOI - PMC - PubMed
Publication types
LinkOut - more resources
Full Text Sources
Other Literature Sources

