Gene Families With Stochastic Exclusive Gene Choice Underlie Cell Adhesion in Mammalian Cells
- PMID: 33996799
- PMCID: PMC8117012
- DOI: 10.3389/fcell.2021.642212
Gene Families With Stochastic Exclusive Gene Choice Underlie Cell Adhesion in Mammalian Cells
Abstract
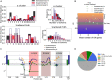
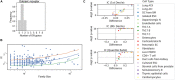
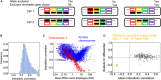
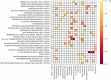
Exclusive stochastic gene choice combines precision with diversity. This regulation enables most T-cells to express exactly one T-cell receptor isoform chosen from a large repertoire, and to react precisely against diverse antigens. Some cells express two receptor isoforms, revealing the stochastic nature of this process. A similar regulation of odorant receptors and protocadherins enable cells to recognize odors and confer individuality to cells in neuronal interaction networks, respectively. We explored whether genes in other families are expressed exclusively by analyzing single-cell RNA-seq data with a simple metric. This metric can detect exclusivity independently of the mean value and the monoallelic nature of gene expression. Chromosomal segments and gene families are more likely to express genes concurrently than exclusively, possibly due to the evolutionary and biophysical aspects of shared regulation. Nonetheless, gene families with exclusive gene choice were detected in multiple cell types, most of them are membrane proteins involved in ion transport and cell adhesion, suggesting the coordination of these two functions. Thus, stochastic exclusive expression extends beyond the prototypical families, permitting precision in gene choice to be combined with the diversity of intercellular interactions.
Keywords: Poisson-binomial distribution; allelic exclusion; basigin; carbonic anhydrase; cell identity; mouse; olfactory receptor; single-cell RNA-seq.
Copyright © 2021 Iakovlev, Faravelli and Becskei.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




Similar articles
-
Gene choice in cancer cells is exclusive in ion transport but concurrent in DNA replication.Comput Struct Biotechnol J. 2024 Jun 10;23:2534-2547. doi: 10.1016/j.csbj.2024.06.004. eCollection 2024 Dec. Comput Struct Biotechnol J. 2024. PMID: 38974885 Free PMC article.
-
One neuron-one receptor rule in the mouse olfactory system.Trends Genet. 2004 Dec;20(12):648-53. doi: 10.1016/j.tig.2004.09.006. Trends Genet. 2004. PMID: 15522461 Review.
-
Coordination of olfactory receptor choice with guidance receptor expression and function in olfactory sensory neurons.PLoS Genet. 2018 Jan 31;14(1):e1007164. doi: 10.1371/journal.pgen.1007164. eCollection 2018 Jan. PLoS Genet. 2018. PMID: 29385124 Free PMC article.
-
An allele-specific, stochastic gene expression process controls the expression of multiple Ly49 family genes and generates a diverse, MHC-specific NK cell receptor repertoire.Eur J Immunol. 1998 Aug;28(8):2407-16. doi: 10.1002/(SICI)1521-4141(199808)28:08<2407::AID-IMMU2407>3.0.CO;2-D. Eur J Immunol. 1998. PMID: 9710218
-
Parallels between Mammalian Mechanisms of Monoallelic Gene Expression.Trends Genet. 2018 Dec;34(12):954-971. doi: 10.1016/j.tig.2018.08.005. Epub 2018 Sep 11. Trends Genet. 2018. PMID: 30217559 Review.
Cited by
-
Decoding core genes and intercellular communication in osteosarcoma: bioinformatic investigation and immune cell profiling for diagnostic and therapeutic insights.Discov Oncol. 2024 Nov 1;15(1):609. doi: 10.1007/s12672-024-01247-y. Discov Oncol. 2024. PMID: 39485636 Free PMC article.
-
Allelic correlation is a marker of trade-offs between barriers to transmission of expression variability and signal responsiveness in genetic networks.Cell Syst. 2022 Dec 21;13(12):1016-1032.e6. doi: 10.1016/j.cels.2022.10.008. Epub 2022 Nov 29. Cell Syst. 2022. PMID: 36450286 Free PMC article.
-
Variability of the innate immune response is globally constrained by transcriptional bursting.Front Mol Biosci. 2023 Jun 27;10:1176107. doi: 10.3389/fmolb.2023.1176107. eCollection 2023. Front Mol Biosci. 2023. PMID: 37441161 Free PMC article.
-
May the Odds Be Ever in Your Favor: Non-deterministic Mechanisms Diversifying Cell Surface Molecule Expression.Front Cell Dev Biol. 2022 Jan 11;9:720798. doi: 10.3389/fcell.2021.720798. eCollection 2021. Front Cell Dev Biol. 2022. PMID: 35087825 Free PMC article. Review.
-
The life and death of RNA across temperatures.Comput Struct Biotechnol J. 2022 Aug 8;20:4325-4336. doi: 10.1016/j.csbj.2022.08.008. eCollection 2022. Comput Struct Biotechnol J. 2022. PMID: 36051884 Free PMC article. Review.
References
-
- Alexander R. A. (1990). A note on averaging correlations. Bull. Psychon. Soc. 28 335–336. 10.3758/bf03334037 - DOI
-
- Almenar-Queralt A., Merkurjev D., Kim H. S., Navarro M., Ma Q., Chaves R. S., et al. (2019). Chromatin establishes an immature version of neuronal protocadherin selection during the naive-to-primed conversion of pluripotent stem cells. Nat. Genet. 51 1691–1701. 10.1038/s41588-019-0526-4 - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources

