Multiplexed detection and differentiation of bacterial enzymes and bacteria by color-encoded sensor hydrogels
- PMID: 33997506
- PMCID: PMC8105640
- DOI: 10.1016/j.bioactmat.2021.04.022
Multiplexed detection and differentiation of bacterial enzymes and bacteria by color-encoded sensor hydrogels
Abstract
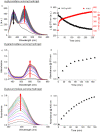
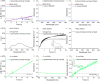
We report on the fabrication and characterization of color-encoded chitosan hydrogels for the rapid, sensitive and specific detection of bacterial enzymes as well as the selective detection of a set of tested bacteria through characteristic enzyme reactions. These patterned sensor hydrogels are functionalized with three different colorimetric enzyme substrates affording the multiplexed detection and differentiation of α-glucosidase, β-galactosidase and β-glucuronidase. The limits of detection of the hydrogels for an observation time of 60 min using a conventional microplate reader correspond to concentrations of 0.2, 3.4 and 4.5 nM of these enzymes, respectively. Based on their different enzyme expression patterns, Staphylococcus aureus strain RN4220, methicillin-resistant S. aureus (MRSA) strain N315, both producing α-glucosidase, but not β-glucuronidase and β-galactosidase, Escherichia coli strain DH5α, producing β-glucuronidase and α-glucosidase, but not β-galactosidase, and the enterohemorrhagic E. coli (EHEC) strain E32511, producing β-galactosidase, but none of the other two enzymes, can be reliably and rapidly distinguished from each other. These results confirm the applicability of enzyme sensing hydrogels for the detection and discrimination of specific enzymes to facilitate differentiation of bacterial strains. Patterned hydrogels thus possess the potential to be further refined as detection units of a multiplexed format to identify certain bacteria for future application in point-of-care microbiological diagnostics in food safety and medical settings.
Keywords: Bacteria detection; Bacterial enzyme; Colorimetric substrates; Multiplexed biosensors; Reporter hydrogels.
© 2021 The Authors.
Conflict of interest statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Figures




References
-
- WHO . 2017. Media Centre. News Release. WHO Publishes List of Bacteria for Which New Antibiotics Are Urgently Needed.http://www.who.int/mediacentre/news/releases/2017/bacteria-antibiotics-n... [cited 2020 Dec.]
-
- Chaired by J. O’NEILL, Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. Government of the United Kingdom; 2016. doi.org/APO-63983.
-
- Le Gall T., Lemercier G., Chevreux S., Tücking K.S., Ravel J., Thétiot F., Jonas U., Schönherr H., Montier T. Ruthenium(II) polypyridyl complexes as photosensitizers for antibacterial photodynamic therapy: a structure-activity study on clinical bacterial strains. ChemMedChem. 2018;13:2229–2239. doi: 10.1002/cmdc.201800392. - DOI - PubMed
-
- Culp E.J., Waglechner N., Wang W., Fiebig-Comyn A.A., Hsu Y.P., Koteva K., Sychantha D., Coombes B.K., Van Nieuwenhze M.S., Brun Y.V., Wright G.D. Evolution-guided discovery of antibiotics that inhibit peptidoglycan remodelling. Nature. 2020;578:582–587. doi: 10.1038/s41586-020-1990-9. - DOI - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

