FYCO1 Regulates Cardiomyocyte Autophagy and Prevents Heart Failure Due to Pressure Overload In Vivo
- PMID: 33997522
- PMCID: PMC8093479
- DOI: 10.1016/j.jacbts.2021.01.001
FYCO1 Regulates Cardiomyocyte Autophagy and Prevents Heart Failure Due to Pressure Overload In Vivo
Abstract
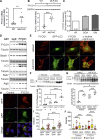
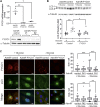
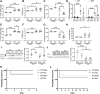
Autophagy is a cellular degradation process that has been implicated in diverse disease processes. The authors provide evidence that FYCO1, a component of the autophagic machinery, is essential for adaptation to cardiac stress. Although the absence of FYCO1 does not affect basal autophagy in isolated cardiomyocytes, it abolishes induction of autophagy after glucose deprivation. Likewise, Fyco1-deficient mice subjected to starvation or pressure overload are unable to respond with induction of autophagy and develop impaired cardiac function. FYCO1 overexpression leads to induction of autophagy in isolated cardiomyocytes and transgenic mouse hearts, thereby rescuing cardiac dysfunction in response to biomechanical stress.
Keywords: BFA, bafilomycin A1; CSA, cell surface area; FYCO1; GFP, green fluorescent protein; KO, knockout; MHC, myosin heavy chain; NRCM, neonatal rat cardiomyocytes; RFP, red fluorescent protein; TAC, transverse aortic constriction; TG, transgenic; WT, wild-type; autophagy; heart failure; mRNA, messenger ribonucleic acid; microRNA, micro–ribonucleic acid.
© 2021 The Authors.
Conflict of interest statement
The authors have reported that they have no relationships relevant to the contents of this paper to disclose.
Figures





Comment in
-
A Novel Inducer of Autophagy in the Heart.JACC Basic Transl Sci. 2021 Apr 27;6(4):381-383. doi: 10.1016/j.jacbts.2021.02.007. eCollection 2021 Apr. JACC Basic Transl Sci. 2021. PMID: 33999043 Free PMC article.
Similar articles
-
Endolysosomal two-pore channels regulate autophagy in cardiomyocytes.J Physiol. 2016 Jun 1;594(11):3061-77. doi: 10.1113/JP271332. Epub 2016 Feb 4. J Physiol. 2016. PMID: 26757341 Free PMC article.
-
Irisin alleviates pressure overload-induced cardiac hypertrophy by inducing protective autophagy via mTOR-independent activation of the AMPK-ULK1 pathway.J Mol Cell Cardiol. 2018 Aug;121:242-255. doi: 10.1016/j.yjmcc.2018.07.250. Epub 2018 Jul 24. J Mol Cell Cardiol. 2018. PMID: 30053525
-
Cardiomyocyte specific overexpression of a 37 amino acid domain of regulator of G protein signalling 2 inhibits cardiac hypertrophy and improves function in response to pressure overload in mice.J Mol Cell Cardiol. 2017 Jul;108:194-202. doi: 10.1016/j.yjmcc.2017.06.007. Epub 2017 Jun 19. J Mol Cell Cardiol. 2017. PMID: 28641980
-
Systemic heme oxygenase-1 transgenic overexpression aggravates pressure overload-induced cardiac hypertrophy in mice.Cell Physiol Biochem. 2011;28(1):25-32. doi: 10.1159/000331710. Epub 2011 Aug 16. Cell Physiol Biochem. 2011. PMID: 21865845
-
Ca2+/calmodulin-dependent protein kinase II is essential in hyperacute pressure overload.J Mol Cell Cardiol. 2020 Jan;138:212-221. doi: 10.1016/j.yjmcc.2019.12.002. Epub 2019 Dec 10. J Mol Cell Cardiol. 2020. PMID: 31836540
Cited by
-
A Novel Inducer of Autophagy in the Heart.JACC Basic Transl Sci. 2021 Apr 27;6(4):381-383. doi: 10.1016/j.jacbts.2021.02.007. eCollection 2021 Apr. JACC Basic Transl Sci. 2021. PMID: 33999043 Free PMC article.
-
The role of FYCO1-dependent autophagy in lens fiber cell differentiation.Autophagy. 2022 Sep;18(9):2198-2215. doi: 10.1080/15548627.2022.2025570. Epub 2022 Mar 28. Autophagy. 2022. PMID: 35343376 Free PMC article.
-
Nicotinamide riboside kinase-2 regulates metabolic adaptation in the ischemic heart.J Mol Med (Berl). 2023 Mar;101(3):311-326. doi: 10.1007/s00109-023-02296-6. Epub 2023 Feb 19. J Mol Med (Berl). 2023. PMID: 36808555
-
The role of the Hippo pathway in autophagy in the heart.Cardiovasc Res. 2023 Jan 18;118(17):3320-3330. doi: 10.1093/cvr/cvac014. Cardiovasc Res. 2023. PMID: 35150237 Free PMC article. Review.
-
Effects of Vitamin D3 Supplementation and Aerobic Training on Autophagy Signaling Proteins in a Rat Model Type 2 Diabetes Induced by High-Fat Diet and Streptozotocin.Nutrients. 2023 Sep 17;15(18):4024. doi: 10.3390/nu15184024. Nutrients. 2023. PMID: 37764807 Free PMC article.
References
-
- Mizushima N., Klionsky D.J. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;27:19–40. - PubMed
-
- Mizushima N., Komatsu M. Autophagy: renovation of cells and tissues. Cell. 2011;147:728–741. - PubMed
-
- Will R.D., Eden M., Just S. Myomasp/LRRC39, a heart- and muscle-specific protein, is a novel component of the sarcomeric M-band and is involved in stretch sensing. Circ Res. 2010;107:1253–1264. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous

