Inter-organelle interactions between the ER and mitotic spindle facilitates Zika protease cleavage of human Kinesin-5 and results in mitotic defects
- PMID: 33997675
- PMCID: PMC8100630
- DOI: 10.1016/j.isci.2021.102385
Inter-organelle interactions between the ER and mitotic spindle facilitates Zika protease cleavage of human Kinesin-5 and results in mitotic defects
Abstract
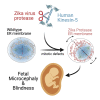
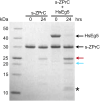
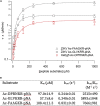
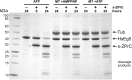
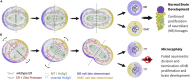
Here we identify human Kinesin-5, Kif11/HsEg5, as a cellular target of Zika protease. We show that Zika NS2B-NS3 protease targets several sites within the motor domain of HsEg5 irrespective of motor binding to microtubules. The native integral ER-membrane protease triggers mitotic spindle positioning defects and a prolonged metaphase delay in cultured cells. Our data support a model whereby loss of function of HsEg5 is mediated by Zika protease and is spatially restricted to the ER-mitotic spindle interface during mitosis. The resulting phenotype is distinct from the monopolar phenotype that typically results from uniform inhibition of HsEg5 by RNAi or drugs. In addition, our data reveal novel inter-organelle interactions between the mitotic apparatus and the surrounding reticulate ER network. Given that Kif11 is haplo-insufficient in humans, and reduced dosage results in microcephaly, we propose that Zika protease targeting of HsEg5 may be a key event in the etiology of Zika syndrome microcephaly.
Keywords: Cytoskeleton; Developmental Genetics; Virology.
Conflict of interest statement
The authors declare no competing interests.
Figures




Similar articles
-
Rigor-type mutation in the kinesin-related protein HsEg5 changes its subcellular localization and induces microtubule bundling.Cell Motil Cytoskeleton. 1998;40(2):174-82. doi: 10.1002/(SICI)1097-0169(1998)40:2<174::AID-CM6>3.0.CO;2-F. Cell Motil Cytoskeleton. 1998. PMID: 9634214
-
The spindle kinesin-like protein HsEg5 is an autoantigen in systemic lupus erythematosus.Arthritis Rheum. 1996 Oct;39(10):1635-42. doi: 10.1002/art.1780391005. Arthritis Rheum. 1996. PMID: 8843853
-
All-trans-retinoic acid-mediated growth inhibition involves inhibition of human kinesin-related protein HsEg5.J Biol Chem. 1999 Jul 2;274(27):18925-31. doi: 10.1074/jbc.274.27.18925. J Biol Chem. 1999. PMID: 10383390
-
The Structure of the Zika Virus Protease, NS2B/NS3pro.Adv Exp Med Biol. 2018;1062:131-145. doi: 10.1007/978-981-10-8727-1_10. Adv Exp Med Biol. 2018. PMID: 29845530 Review.
-
Exploiting the unique features of Zika and Dengue proteases for inhibitor design.Biochimie. 2019 Nov;166:132-141. doi: 10.1016/j.biochi.2019.05.004. Epub 2019 May 9. Biochimie. 2019. PMID: 31077760 Review.
Cited by
-
Emerging roles of cytoskeletal transport and scaffold systems in human viral propagation.Anim Cells Syst (Seoul). 2024 Oct 21;28(1):506-518. doi: 10.1080/19768354.2024.2418332. eCollection 2024. Anim Cells Syst (Seoul). 2024. PMID: 39439927 Free PMC article. Review.
-
Using machine learning to predict protein-protein interactions between a zombie ant fungus and its carpenter ant host.Sci Rep. 2023 Aug 24;13(1):13821. doi: 10.1038/s41598-023-40764-8. Sci Rep. 2023. PMID: 37620441 Free PMC article.
References
-
- Balikova I., Robson A.G., Holder G.E., Ostergaard P., Mansour S., Moore A.T. Ocular manifestations of microcephaly with or without chorioretinopathy, lymphedema or intellectual disability (MCLID) syndrome associated with mutations in KIF11. Acta Ophthalmol. 2016;94:92–98. doi: 10.1111/aos.12759. - DOI - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

