Variable posttranslational modifications of severe acute respiratory syndrome coronavirus 2 nucleocapsid protein
- PMID: 33997890
- PMCID: PMC8241430
- DOI: 10.1093/glycob/cwab044
Variable posttranslational modifications of severe acute respiratory syndrome coronavirus 2 nucleocapsid protein
Abstract
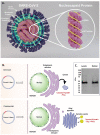
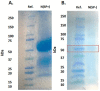
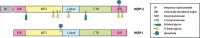
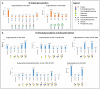
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19), started in 2019 in China and quickly spread into a global pandemic. Nucleocapsid protein (N protein) is highly conserved and is the most abundant protein in coronaviruses and is thus a potential target for both vaccine and point-of-care diagnostics. N Protein has been suggested in the literature as having posttranslational modifications (PTMs), and accurately defining these PTMs is critical for its potential use in medicine. Reports of phosphorylation of N protein have failed to provide detailed site-specific information. We have performed comprehensive glycomics, glycoproteomics and proteomics experiments on two different N protein preparations. Both were expressed in HEK293 cells; one was in-house expressed and purified without a signal peptide (SP) sequence, and the other was commercially produced with a SP channeling it through the secretory pathway. Our results show completely different PTMs on the two N protein preparations. The commercial product contained extensive N- and O-linked glycosylation as well as O-phosphorylation on site Thr393. Conversely, the native N Protein model had O-phosphorylation at Ser176 and no glycosylation, highlighting the importance of knowing the provenance of any commercial protein to be used for scientific or clinical studies. Recent studies have indicated that N protein can serve as an important diagnostic marker for COVID-19 and as a major immunogen by priming protective immune responses. Thus, detailed structural characterization of N protein may provide useful insights for understanding the roles of PTMs on viral pathogenesis, vaccine design and development of point-of-care diagnostics.
Keywords: N protein phosphorylation; N protein site-mapping; SARS-CoV-2 nucleocapsid posttranslational modifications; SARS-CoV-2 phosphoproteomics; glycosylation of SARS-CoV-2 N protein.
© The Author(s) 2021. Published by Oxford University Press. All rights reserved. For permissions, please e-mail: journals.permissions@oup.com.
Figures

Similar articles
-
The SARS-CoV-2 Nucleocapsid Protein and Its Role in Viral Structure, Biological Functions, and a Potential Target for Drug or Vaccine Mitigation.Viruses. 2021 Jun 10;13(6):1115. doi: 10.3390/v13061115. Viruses. 2021. PMID: 34200602 Free PMC article. Review.
-
In-vitro acetylation of SARS-CoV and SARS-CoV-2 nucleocapsid proteins by human PCAF and GCN5.Biochem Biophys Res Commun. 2021 Jun 11;557:273-279. doi: 10.1016/j.bbrc.2021.03.173. Epub 2021 Apr 8. Biochem Biophys Res Commun. 2021. PMID: 33894414 Free PMC article.
-
Evolutionary dynamics of SARS-CoV-2 nucleocapsid protein and its consequences.J Med Virol. 2021 Apr;93(4):2177-2195. doi: 10.1002/jmv.26626. Epub 2020 Nov 10. J Med Virol. 2021. PMID: 33095454
-
Immunoinformatic Analysis of SARS-CoV-2 Nucleocapsid Protein and Identification of COVID-19 Vaccine Targets.Front Immunol. 2020 Oct 28;11:587615. doi: 10.3389/fimmu.2020.587615. eCollection 2020. Front Immunol. 2020. PMID: 33193414 Free PMC article.
-
Protein post-translational modification in SARS-CoV-2 and host interaction.Front Immunol. 2023 Jan 13;13:1068449. doi: 10.3389/fimmu.2022.1068449. eCollection 2022. Front Immunol. 2023. PMID: 36713387 Free PMC article. Review.
Cited by
-
Multi-OMICs landscape of SARS-CoV-2-induced host responses in human lung epithelial cells.iScience. 2023 Jan 20;26(1):105895. doi: 10.1016/j.isci.2022.105895. Epub 2022 Dec 28. iScience. 2023. PMID: 36590899 Free PMC article.
-
Afobazole: a potential drug candidate which can inhibit SARS CoV-2 and mimicry of the human respiratory pacemaker protein.In Silico Pharmacol. 2025 Feb 17;13(1):30. doi: 10.1007/s40203-025-00316-6. eCollection 2025. In Silico Pharmacol. 2025. PMID: 39974371
-
Proteomics-based mass spectrometry profiling of SARS-CoV-2 infection from human nasopharyngeal samples.Mass Spectrom Rev. 2024 Jan-Feb;43(1):193-229. doi: 10.1002/mas.21813. Epub 2022 Sep 29. Mass Spectrom Rev. 2024. PMID: 36177493 Free PMC article. Review.
-
Production and Characterization of a SARS-CoV-2 Nucleocapsid Protein Reference Material.ACS Meas Sci Au. 2022 Sep 2;2(6):620-628. doi: 10.1021/acsmeasuresciau.2c00050. eCollection 2022 Dec 21. ACS Meas Sci Au. 2022. PMID: 36785774 Free PMC article.
-
Mapping the immunopeptidome of seven SARS-CoV-2 antigens across common HLA haplotypes.Nat Commun. 2024 Aug 30;15(1):7547. doi: 10.1038/s41467-024-51959-6. Nat Commun. 2024. PMID: 39214998 Free PMC article.
References
-
- Centers for Disease Control and Prevention . 2020. https://www.cdc.gov/coronavirus/2019-ncov/cases-updates/cases-in-us.html.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

