The quorum-sensing systems of Vibrio campbellii DS40M4 and BB120 are genetically and functionally distinct
- PMID: 33998118
- PMCID: PMC8458232
- DOI: 10.1111/1462-2920.15602
The quorum-sensing systems of Vibrio campbellii DS40M4 and BB120 are genetically and functionally distinct
Abstract
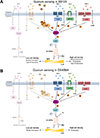
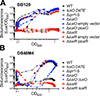
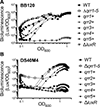
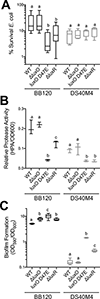
Vibrio campbellii BB120 (previously classified as Vibrio harveyi) is a fundamental model strain for studying quorum sensing in vibrios. A phylogenetic evaluation of sequenced Vibrio strains in Genbank revealed that BB120 is closely related to the environmental isolate V. campbellii DS40M4. We exploited DS40M4's competence for exogenous DNA uptake to rapidly generate greater than 30 isogenic strains with deletions of genes encoding BB120 quorum-sensing system homologues. Our results show that the quorum-sensing circuit of DS40M4 is distinct from BB120 in three ways: (i) DS40M4 does not produce an acyl homoserine lactone autoinducer but encodes an active orphan LuxN receptor, (ii) the quorum regulatory small RNAs (Qrrs) are not solely regulated by autoinducer signalling through the response regulator LuxO and (iii) the DS40M4 quorum-sensing regulon is much smaller than BB120 (~100 genes vs. ~400 genes, respectively). Using comparative genomics to expand our understanding of quorum-sensing circuit diversity, we observe that conservation of LuxM/LuxN proteins differs widely both between and within Vibrio species. These strains are also phenotypically distinct: DS40M4 exhibits stronger interbacterial cell killing, whereas BB120 forms more robust biofilms and is bioluminescent. These results underscore the need to examine wild isolates for a broader view of bacterial diversity in the marine ecosystem.
© 2021 Society for Applied Microbiology and John Wiley & Sons Ltd.
Conflict of interest statement
Conflict of Interest Statement
The authors declare that they have no conflicts of interest.
Figures



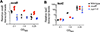
References
-
- Bassler BL, Wright M, Showalter RE, and Silverman MR (1993) Intercellular signalling in Vibrio harveyi: sequence and function of genes regulating expression of luminescence. Mol Microbiol 9: 773–786. - PubMed
Publication types
MeSH terms
Substances
Supplementary concepts
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

