Transcriptome-wide association analysis identifies DACH1 as a kidney disease risk gene that contributes to fibrosis
- PMID: 33998598
- PMCID: PMC8121513
- DOI: 10.1172/JCI141801
Transcriptome-wide association analysis identifies DACH1 as a kidney disease risk gene that contributes to fibrosis
Abstract
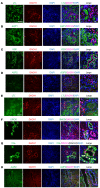
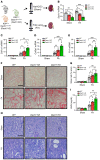
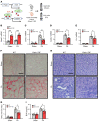
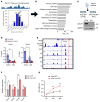
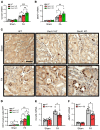
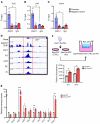
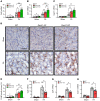
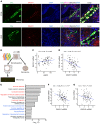
Genome-wide association studies (GWAS) for kidney function identified hundreds of risk regions; however, the causal variants, target genes, cell types, and disease mechanisms remain poorly understood. Here, we performed transcriptome-wide association studies (TWAS), summary Mendelian randomization, and MetaXcan to identify genes whose expression mediates the genotype effect on the phenotype. Our analyses identified Dachshund homolog 1 (DACH1), a cell-fate determination factor. GWAS risk variant was associated with lower DACH1 expression in human kidney tubules. Human and mouse kidney single-cell open chromatin data (snATAC-Seq) prioritized estimated glomerular filtration rate (eGFR) GWAS variants located on an intronic regulatory region in distal convoluted tubule cells. CRISPR-Cas9-mediated gene editing confirmed the role of risk variants in regulating DACH1 expression. Mice with tubule-specific Dach1 deletion developed more severe renal fibrosis both in folic acid and diabetic kidney injury models. Mice with tubule-specific Dach1 overexpression were protected from folic acid nephropathy. Single-cell RNA sequencing, chromatin immunoprecipitation, and functional analysis indicated that DACH1 controls the expression of cell cycle and myeloid chemotactic factors, contributing to macrophage infiltration and fibrosis development. In summary, integration of GWAS, TWAS, single-cell epigenome, expression analyses, gene editing, and functional validation in different mouse kidney disease models identified DACH1 as a kidney disease risk gene.
Keywords: Chronic kidney disease; Genetic variation; Genetics; Nephrology.
Conflict of interest statement
Figures




Comment in
-
DACH1 as a multifaceted and potentially druggable susceptibility factor for kidney disease.J Clin Invest. 2021 May 17;131(10):e149043. doi: 10.1172/JCI149043. J Clin Invest. 2021. PMID: 33998596 Free PMC article.
-
DACH1 protects podocytes and tubules from injury.Nat Rev Nephrol. 2021 Aug;17(8):511. doi: 10.1038/s41581-021-00450-2. Nat Rev Nephrol. 2021. PMID: 34083770 No abstract available.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

